basecount
Count nucleotides in sequence
Description
Examples
Count Nucleotides in Sequence
Count the bases in a DNA sequence and return the results in a structure.
bases = basecount('TAGCTGGCCAAGCGAGCTTG')bases = struct with fields:
A: 4
C: 5
G: 7
T: 4
Get the number of adenosine (A) bases.
bases.A
ans = 4
Create a bar graph comparing the number of each nucleotide.
basecount('TAGCTGGCCAAGCGAGCTTG',Chart="bar")
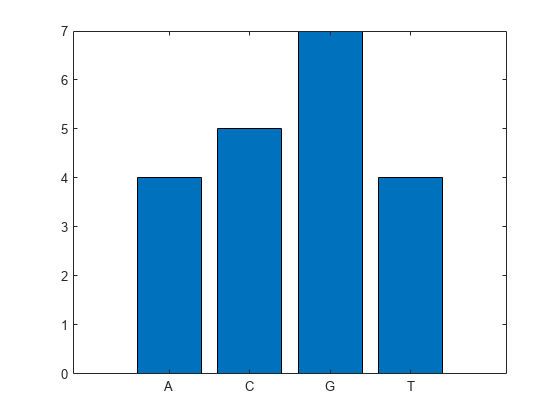
ans = struct with fields:
A: 4
C: 5
G: 7
T: 4
Count the bases in a DNA sequence containing ambiguous characters (R, Y, K, M, S, W, B, D, H, V, or N), listing each of them in a separate field.
basecount('ABCDGGCCAAGCGAGCTTG',Ambiguous="individual")
ans = struct with fields:
A: 4
C: 5
G: 6
T: 2
R: 0
Y: 0
K: 0
M: 0
S: 0
W: 0
B: 1
D: 1
H: 0
V: 0
N: 0
Input Arguments
SeqNT — Nucleotide sequence
character vector | string scalar | row vector of integers | structure
Nucleotide sequence, specified as one of the following.
Character vector or string scalar consisting of the characters
A,C,G,T, andU, and ambiguous charactersR,Y,K,M,S,W,B,D,H,V, andN.Row vector of integers specifying a nucleotide sequence. For information on valid integers, see Mapping Nucleotide Integers to Letter Codes.
Structure that contains a nucleotide sequence in the
Sequencefield. Thefastaread,fastqread,emblread,getembl,genbankread, andgetgenbankfunctions return structures with aSequencefield.
Example: NTStruct = basecount('CGACTT') counts the number of times
of each nucleotide occurs in the sequence.
Data Types: double | char | string | struct
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: NTStruct =
basecount("ACGGTC",Ambiguous="individual")
Ambiguous — Method for counting ambiguous nucleotide characters
"ignore" (default) | "bundle" | "prorate" | "individual" | "warn"
Method for counting ambiguous nucleotide characters (R,
Y, K, M,
S, W, B,
D, H, V, and
N), specified as one of the following.
"ignore"—basecountskips ambiguous characters."bundle"—basecountcounts ambiguous characters and reports the total count in theAmbiguousfield."prorate"—basecountcounts ambiguous characters and distributes the total number evenly between all possible unambiguous nucleotide fields. For example, the count for the characterRis distributed evenly between theAandGfields."individual"—basecountcounts ambiguous characters and reports them in individual fields."warn"—basecountskips ambiguous characters and displays a warning.
Example: NTStruct = basecount("CGRTTMSA",Ambiguous="bundle")
reports the total number of ambiguous characters in the Ambiguous
field of NTStruct.
Data Types: char | string
Gaps — Flag to count or ignore gaps
false (default) | true
Flag to count or ignore gaps, specified as true or
false. Gaps are indicated by a hyphen
(-).
If you set this option to true, then
basecount counts the gaps and reports the total count in the
Gaps field.
Data Types: logical
Chart — Type of chart
"pie" | "bar"
Type of chart to display the proportions of nucleotides, specified as
"pie" or "bar".
Data Types: char | string
Output Arguments
NTStruct — Nucleotide counts
structure
Nucleotide counts, returned as a structure containing the fields
A, C, G, and
T. Uracil nucleotides (U) are added to the
T field. Additional fields can be present, depending on the value
of Ambiguous and Gaps.
Version History
Introduced before R2006a
See Also
aacount | baselookup | codoncount | cpgisland | dimercount | nmercount | ntdensity | seqviewer
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)