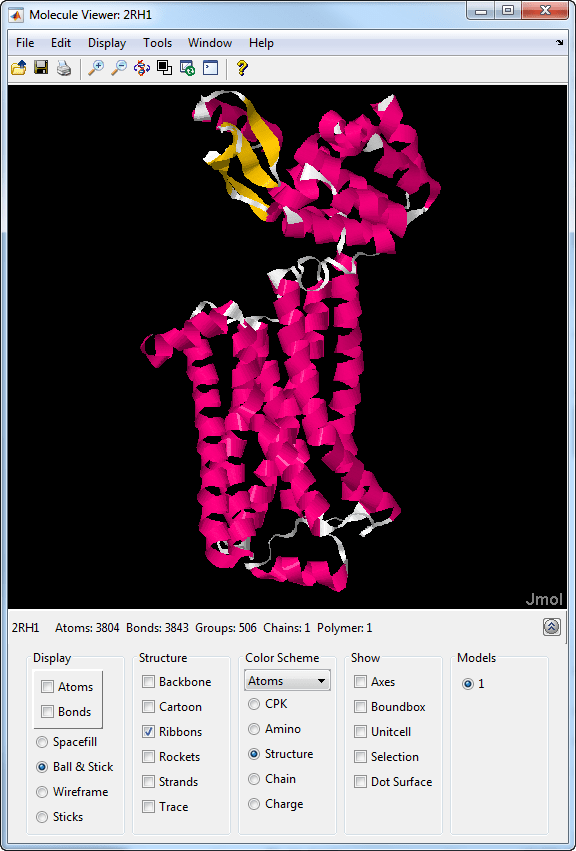
Molecule Viewer
(To be removed) Display and manipulate 3-D molecule structure
The app will be removed in a future release.
Description
The Molecule Viewer app lets you display and manipulate 3-D molecular structures.
You can:
Import structural information directly from the Protein Data Bank (PDB) database or other supported files.
Measure distances and dihedral angles.
Display molecular surfaces, such as van der Waals or solvent-accessible surfaces.
Select different visualization and color schemes to display a molecule, such as the ribbon or backbone representation.
Run RasMol script commands from within the app.
Open the Molecule Viewer App
MATLAB® Toolstrip: On the Apps tab, under Computational Biology, click the app icon.
MATLAB command prompt: Enter
molviewer.