subtree
Extract phylogenetic subtree
Syntax
Tree2 = subtree(Tree1, Nodes)
Description
Tree2 = subtree(Tree1, Nodes)Tree2) where the new root
is the first common ancestor of the Nodes vector
from Tree1. Nodes in the tree are indexed
as [1:NUMLEAVES] for the leaves and as [NUMLEAVES+1:NUMLEAVES+NUMBRANCHES] for
the branches. Nodes can also be a logical array of following sizes [NUMLEAVES+NUMBRANCHES
x 1], [NUMLEAVES x 1] or [NUMBRANCHES
x 1].
Examples
Load a phylogenetic tree created from a protein family.
tr = phytreeread('pf00002.tree');Get the subtree that contains the VIPR2 and GLR human proteins.
sel = getbyname(tr,{'vipr2_human','glr_human'}); sel = any(sel,2); tr = subtree(tr,sel); view(tr)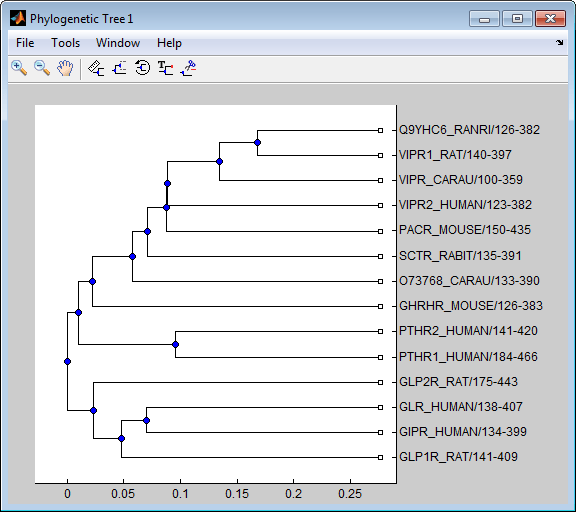
Version History
Introduced before R2006a
