Create Model of Receptor-Ligand Kinetics
This example shows how to create and simulate a simple model of receptor-ligand kinetics using the SimBiology Model Builder and SimBiology Model Analyzer apps.
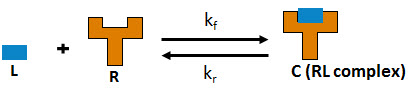
In this model, ligand L and receptor R species form receptor-ligand complexes through reversible binding reactions. These reactions are defined using mass action kinetics by , where kf and kr are forward and reverse rate constants. L, R, and C are the concentrations of the ligand, receptor, and receptor-ligand complex, respectively.
Open Model Builder App
Click the SimBiology Model Builder icon on the
Apps tab or enter simBiologyModelBuilder at the command
line.
Build Model
On the Home tab of the app, select Model > Create New Blank Model. Enter
m1as the name for the model. The app creates an empty compartmentunnamedand displays the compartment on the Diagram tab.Drag and drop three species blocks
 and one reaction block
and one reaction block  into the compartment. Optionally, you can rename the species and compartment by double-clicking the default names. For instance, change
into the compartment. Optionally, you can rename the species and compartment by double-clicking the default names. For instance, change unnamedtocell.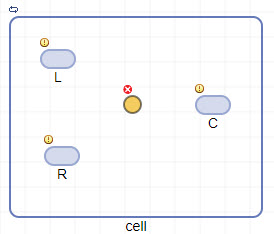
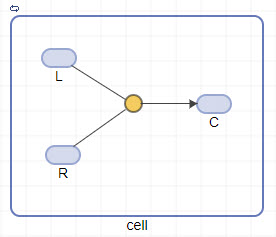
To connect the species to the reaction, press and hold the Ctrl key (on Windows® and Linux®) or the Option key (on macOS), click the species block, and drag the line.
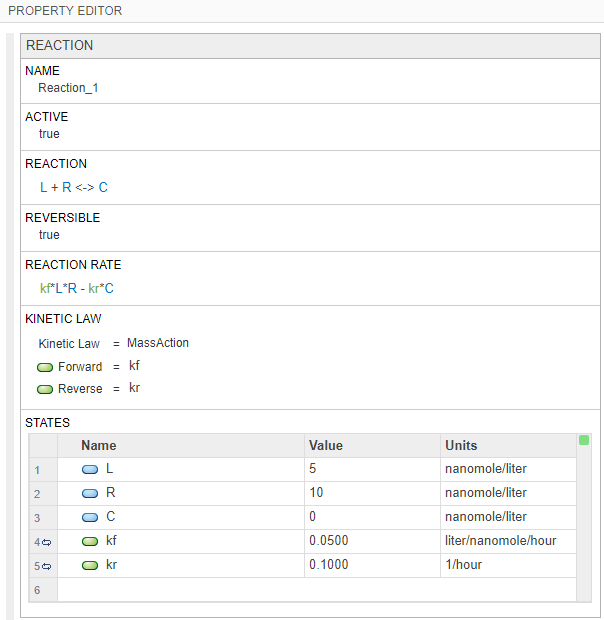
Click the reaction block to see its properties in the Property Editor pane. Set the following parameters.
Select Reversible > true.
In the States table, update the values of L to
5and R to10. Set the units of the L, R, and C species tonanomole/liter.Set the value of the forward rate parameter kf to
0.05. Set the unit toliter/nanomole/hour.Set the value of the reverse rate parameter kr to
0.1with the unit1/hour.
Click the cell compartment in the diagram. In the Property Editor pane, set its Units to liter.
Simulate Model
On the Home tab, click the Model Analyzer icon to open the SimBiology Model Analyzer app.
In the Model Analyzer app, select Program > Simulate Model on the Home tab. The Program1 tab opens.
In the Simulation step of the program, set the Stop Time to 20 seconds because the model reaches a saturated state after that.
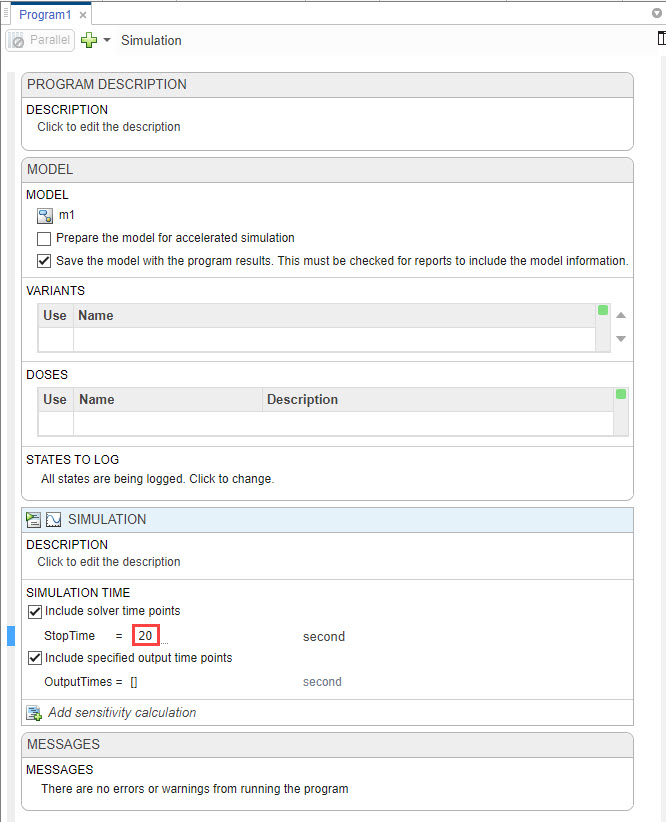
Click Run from the Home tab.
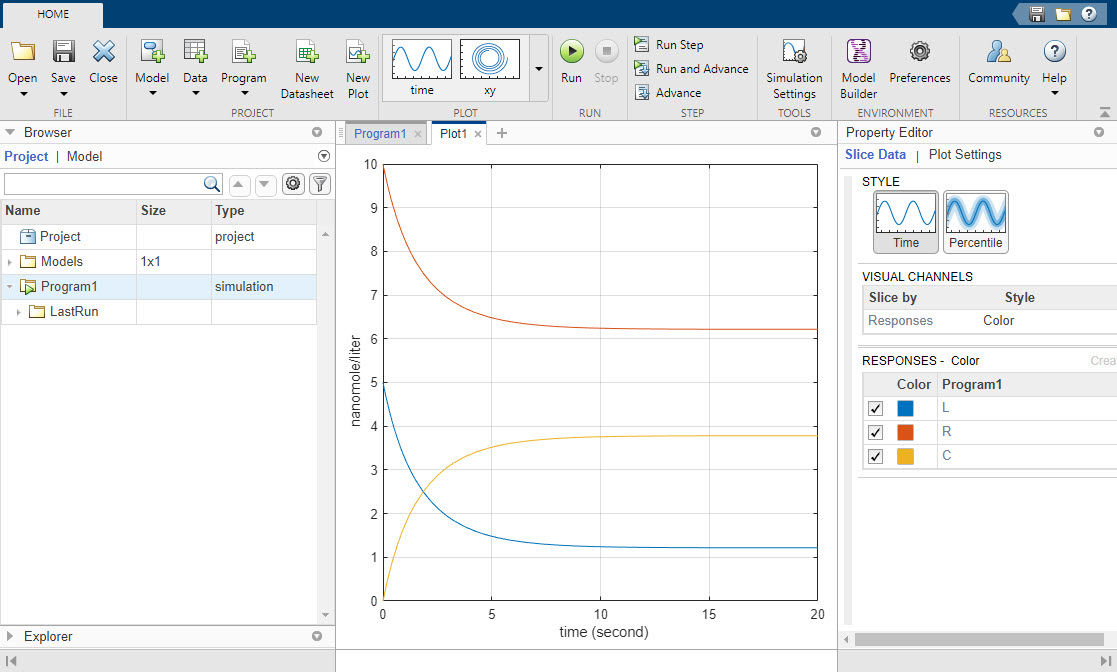
Running the program plots the results in the Plot1 tab. The plot shows the simulated responses in different colors. The program stores the simulation results in the LastRun folder of the program.
See Also
SimBiology Model Builder | SimBiology Model Analyzer
Related Topics
- SimBiology Apps
- Calculate NCA Parameters and Fit Model to PK/PD Data Using SimBiology Model Analyzer
- Find Important Tumor Growth Parameters with Local Sensitivity Analysis Using SimBiology Model Analyzer
- Explore Biological Variability with Virtual Patients Using SimBiology Model Analyzer
- Scan Dosing Regimens Using SimBiology Model Analyzer App
