predict
Predict labels using classification ensemble model
Description
labels = predict(ens,X,Name=Value)
[ also returns a matrix of classification scores indicating the likelihood that a label comes from a particular
class, using any of the input argument combinations in the previous syntaxes. For
each observation in labels,scores]
= predict(___)X, the predicted class label corresponds to
the maximum score among all classes.
Examples
Predict Class Labels Using Classification Ensemble
Load Fisher's iris data set. Determine the sample size.
load fisheriris
N = size(meas,1);Partition the data into training and test sets. Hold out 10% of the data for testing.
rng(1); % For reproducibility cvp = cvpartition(N,'Holdout',0.1); idxTrn = training(cvp); % Training set indices idxTest = test(cvp); % Test set indices
Store the training data in a table.
tblTrn = array2table(meas(idxTrn,:)); tblTrn.Y = species(idxTrn);
Train a classification ensemble using AdaBoostM2 and the training set. Specify tree stumps as the weak learners.
t = templateTree('MaxNumSplits',1); Mdl = fitcensemble(tblTrn,'Y','Method','AdaBoostM2','Learners',t);
Predict labels for the test set. You trained model using a table of data, but you can predict labels using a matrix.
labels = predict(Mdl,meas(idxTest,:));
Construct a confusion matrix for the test set.
confusionchart(species(idxTest),labels)
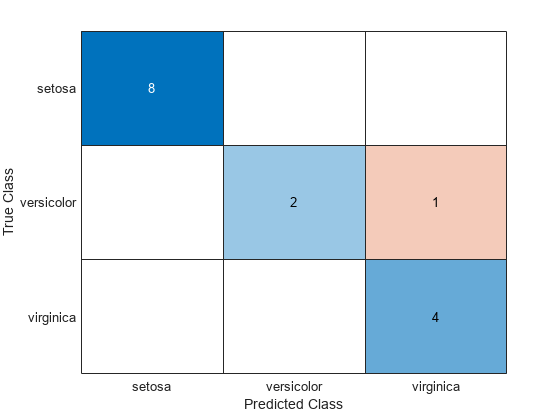
Mdl misclassifies one versicolor iris as virginica in the test set.
Assess Performance of Ensemble of Boosted Trees
Create an ensemble of boosted trees and inspect the importance of each predictor. Using test data, assess the classification accuracy of the ensemble.
Load the arrhythmia data set. Determine the class representations in the data.
load arrhythmia
Y = categorical(Y);
tabulate(Y) Value Count Percent
1 245 54.20%
2 44 9.73%
3 15 3.32%
4 15 3.32%
5 13 2.88%
6 25 5.53%
7 3 0.66%
8 2 0.44%
9 9 1.99%
10 50 11.06%
14 4 0.88%
15 5 1.11%
16 22 4.87%
The data set contains 16 classes, but not all classes are represented (for example, class 13). Most observations are classified as not having arrhythmia (class 1). The data set is highly discrete with imbalanced classes.
Combine all observations with arrhythmia (classes 2 through 15) into one class. Remove those observations with an unknown arrhythmia status (class 16) from the data set.
idx = (Y ~= "16"); Y = Y(idx); X = X(idx,:); Y(Y ~= "1") = "WithArrhythmia"; Y(Y == "1") = "NoArrhythmia"; Y = removecats(Y);
Create a partition that evenly splits the data into training and test sets.
rng("default") % For reproducibility cvp = cvpartition(Y,"Holdout",0.5); idxTrain = training(cvp); idxTest = test(cvp);
cvp is a cross-validation partition object that specifies the training and test sets.
Train an ensemble of 100 boosted classification trees using AdaBoostM1. Specify to use tree stumps as the weak learners. Also, because the data set contains missing values, specify to use surrogate splits.
t = templateTree("MaxNumSplits",1,"Surrogate","on"); numTrees = 100; mdl = fitcensemble(X(idxTrain,:),Y(idxTrain),"Method","AdaBoostM1", ... "NumLearningCycles",numTrees,"Learners",t);
mdl is a trained ClassificationEnsemble model.
Inspect the importance measure for each predictor.
predImportance = predictorImportance(mdl); bar(predImportance) title("Predictor Importance") xlabel("Predictor") ylabel("Importance Measure")
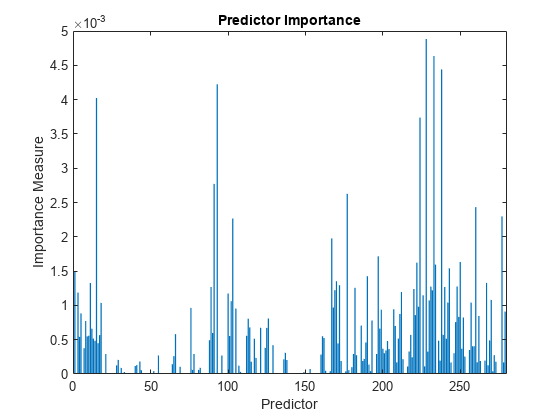
Identify the top ten predictors in terms of their importance.
[~,idxSort] = sort(predImportance,"descend");
idx10 = idxSort(1:10)idx10 = 1×10
228 233 238 93 15 224 91 177 260 277
Classify the test set observations. View the results using a confusion matrix. Blue values indicate correct classifications, and red values indicate misclassified observations.
predictedValues = predict(mdl,X(idxTest,:)); confusionchart(Y(idxTest),predictedValues)

Compute the accuracy of the model on the test data.
error = loss(mdl,X(idxTest,:),Y(idxTest), ... "LossFun","classiferror"); accuracy = 1 - error
accuracy = 0.7731
accuracy estimates the fraction of correctly classified observations.
Input Arguments
ens — Classification ensemble model
ClassificationEnsemble model object | CompactClassificationEnsemble model object
Full classification ensemble model, specified as a ClassificationEnsemble model object trained with fitcensemble, or a CompactClassificationEnsemble model object created with compact.
X — Predictor data
numeric matrix | table
Predictor data to be classified, specified as a numeric matrix or a table.
Each row of X corresponds to one observation, and each
column corresponds to one variable.
For a numeric matrix:
The variables that make up the columns of
Xmust have the same order as the predictor variables used to trainens.If you trained
ensusing a table (for example,tbl),Xcan be a numeric matrix iftblcontains only numeric predictor variables. To treat numeric predictors intblas categorical during training, specify categorical predictors using theCategoricalPredictorsname-value argument offitcensemble. Iftblcontains heterogeneous predictor variables (for example, numeric and categorical data types) andXis a numeric matrix,predictissues an error.
For a table:
predictdoes not support multicolumn variables or cell arrays other than cell arrays of character vectors.If you trained
ensusing a table (for example,tbl), all predictor variables inXmust have the same variable names and data types as those used to trainens(stored inens.PredictorNames). However, the column order ofXdoes not need to correspond to the column order oftbl.tblandXcan contain additional variables, such as response variables and observation weights, butpredictignores them.If you trained
ensusing a numeric matrix, then the predictor names inens.PredictorNamesmust be the same as the corresponding predictor variable names inX. To specify predictor names during training, use thePredictorNamesname-value argument offitcensemble. All predictor variables inXmust be numeric vectors.Xcan contain additional variables, such as response variables and observation weights, butpredictignores them.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: predict(ens,X,Learners=[1 2 3 5],UseParallel=true)
specifies to use the first, second, third, and fifth learners in the ensemble
ens, and to perform computations in
parallel.
Learners — Indices of weak learners
[1:ens.NumTrained] (default) | vector of positive integers
Indices of weak learners in the ensemble to use in
predict, specified as a vector of positive integers in the range
[1:ens.NumTrained]. By default, all learners are used.
Example: Learners=[1 2 4]
Data Types: single | double
UseObsForLearner — Option to use observations for learners
true(N,T) (default) | logical matrix
Option to use observations for learners, specified as a logical matrix of size
N-by-T, where:
When UseObsForLearner(i,j) is true (default),
learner j is used in predicting the class of row i
of X.
Example: UseObsForLearner=logical([1 1; 0 1; 1 0])
Data Types: logical matrix
UseParallel — Flag to run in parallel
false or 0 (default) | true or 1
Flag to run in parallel, specified as a numeric or logical
1 (true) or 0
(false). If you specify UseParallel=true, the
predict function executes for-loop iterations by
using parfor. The loop runs in parallel when you
have Parallel Computing Toolbox™.
Example: UseParallel=true
Data Types: logical
Output Arguments
labels — Predicted class labels
categorical array | character array | logical array | numeric array | cell array of character vectors
Predicted class labels, returned as a categorical, character, logical, or
numeric array, or a cell array of character vectors.
labels has the same data type as the labels used to
train ens. (The software treats string arrays as cell arrays of character
vectors.)
The predict function classifies an observation into the class yielding the highest score. For an observation with NaN scores, the
function classifies the observation into the majority class, which makes up the largest
proportion of the training labels.
scores — Class scores
numeric matrix
Class scores, returned as a numeric matrix with one row per observation and one column per class. For each observation and each class, the score represents the confidence that the observation originates from that class. A higher score indicates a higher confidence. For more information, see Score (ensemble).
More About
Score (ensemble)
For ensembles, a classification score represents the confidence that an observation originates from a specific class. The higher the score, the higher the confidence.
Different ensemble algorithms have different definitions for their scores. Furthermore, the range of scores depends on ensemble type. For example:
Bagscores range from0to1. You can interpret these scores as probabilities averaged over all the trees in the ensemble.AdaBoostM1,GentleBoost, andLogitBoostscores range from –∞ to ∞. You can convert these scores to probabilities by setting theScoreTransformproperty ofensto"doublelogit"before passingenstopredict:Alternatively, you can specifyens.ScoreTransform = "doublelogit"; [labels,scores] = predict(ens,X);ScoreTransform="doublelogit"in the call tofitcensemblewhen you createens.
For more information on the different ensemble algorithms and how they compute scores, see Ensemble Algorithms.
Alternative Functionality
Simulink Block
To integrate the prediction of an ensemble into Simulink®, you can use the ClassificationEnsemble Predict block in the Statistics and Machine Learning Toolbox™ library or a MATLAB® Function block with the predict function. For
examples, see Predict Class Labels Using ClassificationEnsemble Predict Block and Predict Class Labels Using MATLAB Function Block.
When deciding which approach to use, consider the following:
If you use the Statistics and Machine Learning Toolbox library block, you can use the Fixed-Point Tool (Fixed-Point Designer) to convert a floating-point model to fixed point.
Support for variable-size arrays must be enabled for a MATLAB Function block with the
predictfunction.If you use a MATLAB Function block, you can use MATLAB functions for preprocessing or post-processing before or after predictions in the same MATLAB Function block.
Extended Capabilities
Tall Arrays
Calculate with arrays that have more rows than fit in memory.
Usage notes and limitations:
You cannot use the
UseParallelname-value argument with tall arrays.
For more information, see Tall Arrays.
C/C++ Code Generation
Generate C and C++ code using MATLAB® Coder™.
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for thepredictfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls thepredictfunction. Then usecodegento generate code for the entry-point function.To generate single-precision C/C++ code for
predict, specify the name-value argument"DataType","single"when you call theloadLearnerForCoderfunction.You can also generate fixed-point C/C++ code for
predict. Fixed-point code generation requires an additional step that defines the fixed-point data types of the variables required for prediction. Create a fixed-point data type structure by using the data type function generated bygenerateLearnerDataTypeFcn, and then use the structure as an input argument ofloadLearnerForCoderin an entry-point function. Generating fixed-point C/C++ code requires MATLAB Coder™ and Fixed-Point Designer™.Generating fixed-point code for
predictincludes propagating data types for individual learners and, therefore, can be time consuming.This table contains notes about the arguments of
predict. Arguments not included in this table are fully supported.Argument Notes and Limitations ensFor the usage notes and limitations of the model object, see Code Generation of the
CompactClassificationEnsembleobject.XFor general code generation,
Xmust be a single-precision or double-precision matrix or a table containing numeric variables, categorical variables, or both.For fixed-point code generation,
Xmust be a fixed-point matrix.The number of rows, or observations, in
Xcan be a variable size, but the number of columns inXmust be fixed.If you want to specify
Xas a table, then your model must be trained using a table, and your entry-point function for prediction must do the following:Accept data as arrays.
Create a table from the data input arguments and specify the variable names in the table.
Pass the table to
predict.
For an example of this table workflow, see Generate Code to Classify Data in Table. For more information on using tables in code generation, see Code Generation for Tables (MATLAB Coder) and Table Limitations for Code Generation (MATLAB Coder).
Name-value arguments Names in name-value arguments must be compile-time constants. For example, to allow user-defined indices up to 5 weak learners in the generated code, include
{coder.Constant('Learners'),coder.typeof(0,[1,5],[0,1])}in the-argsvalue ofcodegen(MATLAB Coder)."Learners"For fixed-point code generation, the
"Learners"value must have an integer data type.
For more information, see Introduction to Code Generation.
Automatic Parallel Support
Accelerate code by automatically running computation in parallel using Parallel Computing Toolbox™.
To run in parallel, set the UseParallel name-value argument to
true in the call to this function.
For more general information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
You cannot use UseParallel with tall or GPU arrays or in code generation.
GPU Arrays
Accelerate code by running on a graphics processing unit (GPU) using Parallel Computing Toolbox™.
Usage notes and limitations:
The
predictfunction does not support ensembles trained using decision tree learners with surrogate splits.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2011a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)