wavemngr
Wavelet manager
Syntax
Description
Use wavemngr to add, delete, restore, or read
wavelets.
wavemngr('add',
adds a wavelet family to the toolbox. These parameters define the wavelet family:FN,FSN,WT,NUMS,FILE)
FN— Family nameFSN— Family short nameWT— Wavelet family typeNUMS— Wavelet parametersFILE— Wavelet definition file
Note
When you use wavemngr to add a wavelet family,
three wavelet extension files are created in the current folder: the two
ASCII files wavelets.asc and
wavelets.prv, and the MAT-file
wavelets.inf. If you add a new wavelet family, it
is available in this folder only.
wavemngr('restore') restores the previous
wavelets.asc ASCII file
wavemngr('restore',IN2) restores the initial
wavelets.asc ASCII file. Here IN2 is a
dummy argument.
Examples
Wavelet Names and Family Names
List the wavelet families available by default.
wavemngr('read')ans = 23x35 char array
'==================================='
'Haar ->->haar '
'Daubechies ->->db '
'Symlets ->->sym '
'Coiflets ->->coif '
'BiorSplines ->->bior '
'ReverseBior ->->rbio '
'Meyer ->->meyr '
'DMeyer ->->dmey '
'Gaussian ->->gaus '
'Mexican_hat ->->mexh '
'Morlet ->->morl '
'Complex Gaussian ->->cgau '
'Shannon ->->shan '
'Frequency B-Spline ->->fbsp '
'Complex Morlet ->->cmor '
'Fejer-Korovkin ->->fk '
'Best-localized Daubechies->->bl '
'Morris minimum-bandwidth ->->mb '
'Beylkin ->->beyl '
'Vaidyanathan ->->vaid '
'Han linear-phase moments ->->han '
'==================================='
List all wavelets.
wavemngr('read',1)ans = 89x44 char array
'=================================== '
'Haar ->->haar '
'=================================== '
'Daubechies ->->db '
'------------------------------ '
'db1->db2->db3->db4-> '
'db5->db6->db7->db8-> '
'db9->db10->db**-> '
'=================================== '
'Symlets ->->sym '
'------------------------------ '
'sym2->sym3->sym4->sym5-> '
'sym6->sym7->sym8->sym**-> '
'=================================== '
'Coiflets ->->coif '
'------------------------------ '
'coif1->coif2->coif3->coif4-> '
'coif5-> '
'=================================== '
'BiorSplines ->->bior '
'------------------------------ '
'bior1.1->bior1.3->bior1.5->bior2.2-> '
'bior2.4->bior2.6->bior2.8->bior3.1-> '
'bior3.3->bior3.5->bior3.7->bior3.9-> '
'bior4.4->bior5.5->bior6.8-> '
'=================================== '
'ReverseBior ->->rbio '
'------------------------------ '
'rbio1.1->rbio1.3->rbio1.5->rbio2.2-> '
'rbio2.4->rbio2.6->rbio2.8->rbio3.1-> '
'rbio3.3->rbio3.5->rbio3.7->rbio3.9-> '
'rbio4.4->rbio5.5->rbio6.8-> '
'=================================== '
'Meyer ->->meyr '
'=================================== '
'DMeyer ->->dmey '
'=================================== '
'Gaussian ->->gaus '
'------------------------------ '
'gaus1->gaus2->gaus3->gaus4-> '
'gaus5->gaus6->gaus7->gaus8-> '
'=================================== '
'Mexican_hat ->->mexh '
'=================================== '
'Morlet ->->morl '
'=================================== '
'Complex Gaussian ->->cgau '
'------------------------------ '
'cgau1->cgau2->cgau3->cgau4-> '
'cgau5->cgau6->cgau7->cgau8-> '
'=================================== '
'Shannon ->->shan '
'------------------------------ '
'shan1-1.5->shan1-1->shan1-0.5->shan1-0.1-> '
'shan2-3->shan**-> '
'=================================== '
'Frequency B-Spline ->->fbsp '
'------------------------------ '
'fbsp1-1-1.5->fbsp1-1-1->fbsp1-1-0.5->fbsp2-1-1->'
'fbsp2-1-0.5->fbsp2-1-0.1->fbsp**-> '
'=================================== '
'Complex Morlet ->->cmor '
'------------------------------ '
'cmor1-1.5->cmor1-1->cmor1-0.5->cmor1-1-> '
'cmor1-0.5->cmor1-0.1->cmor**-> '
'=================================== '
'Fejer-Korovkin ->->fk '
'------------------------------ '
'fk4->fk6->fk8->fk14-> '
'fk18->fk22-> '
'=================================== '
'Best-localized Daubechies->->bl '
'------------------------------ '
'bl7->bl9->bl10-> '
'=================================== '
'Morris minimum-bandwidth ->->mb '
'------------------------------ '
'mb4.2->mb8.2->mb8.3->mb8.4-> '
'mb10.3->mb12.3->mb14.3->mb16.3-> '
'mb18.3->mb24.3->mb32.3-> '
'=================================== '
'Beylkin ->->beyl '
'=================================== '
'Vaidyanathan ->->vaid '
'=================================== '
'Han linear-phase moments ->->han '
'------------------------------ '
'han2.3->han3.3->han4.5->han5.5-> '
'=================================== '
Add Wavelet Families
This example shows how to add new compactly supported orthogonal wavelets to the toolbox. These wavelets, which are a slight generalization of the Daubechies wavelets, are based on the use of Bernstein polynomials and are due to Kateb and Lemarié.
Add a new family of orthogonal wavelets. You must define:
Family Name:
LemarieFamily Short Name:
lemType of wavelet:
1 (orth)Wavelet numbers:
1 2 3 4 5File driver:
lemwavf
The source code for lemwavf.m is in the same folder as this example. The input argument of lemwavf is a character vector of the form lemN, where N = 1, 2, 3, 4, or 5.
wavemngr('add','Lemarie','lem',1,'1 2 3 4 5','lemwavf')
The ASCII file wavelets.asc is saved as wavelets.prv, then information defining the new family is added to wavelets.asc, and the MAT-file wavelets.inf is generated.
Note that wavemngr works on the current folder. If you add a new wavelet family, it is available in this folder only.
List the available wavelet families. Confirm the new wavelet family is added.
wavemngr('read')ans = 24x35 char array
'==================================='
'Haar ->->haar '
'Daubechies ->->db '
'Symlets ->->sym '
'Coiflets ->->coif '
'BiorSplines ->->bior '
'ReverseBior ->->rbio '
'Meyer ->->meyr '
'DMeyer ->->dmey '
'Gaussian ->->gaus '
'Mexican_hat ->->mexh '
'Morlet ->->morl '
'Complex Gaussian ->->cgau '
'Shannon ->->shan '
'Frequency B-Spline ->->fbsp '
'Complex Morlet ->->cmor '
'Fejer-Korovkin ->->fk '
'Best-localized Daubechies->->bl '
'Morris minimum-bandwidth ->->mb '
'Beylkin ->->beyl '
'Vaidyanathan ->->vaid '
'Han linear-phase moments ->->han '
'Lemarie ->->lem '
'==================================='
Remove the added family. Regenerate the list of wavelet families.
wavemngr('del','Lemarie') wavemngr('read')
ans = 23x35 char array
'==================================='
'Haar ->->haar '
'Daubechies ->->db '
'Symlets ->->sym '
'Coiflets ->->coif '
'BiorSplines ->->bior '
'ReverseBior ->->rbio '
'Meyer ->->meyr '
'DMeyer ->->dmey '
'Gaussian ->->gaus '
'Mexican_hat ->->mexh '
'Morlet ->->morl '
'Complex Gaussian ->->cgau '
'Shannon ->->shan '
'Frequency B-Spline ->->fbsp '
'Complex Morlet ->->cmor '
'Fejer-Korovkin ->->fk '
'Best-localized Daubechies->->bl '
'Morris minimum-bandwidth ->->mb '
'Beylkin ->->beyl '
'Vaidyanathan ->->vaid '
'Han linear-phase moments ->->han '
'==================================='
Restore the previous ASCII file wavelets.prv, then build the MAT-file wavelets.inf. List the restored wavelets. Because wavemngr reads the ASCII file in the current working directory, the new family appears in the list.
wavemngr('restore') wavemngr('read',1)
ans = 94x44 char array
'=================================== '
'Haar ->->haar '
'=================================== '
'Daubechies ->->db '
'------------------------------ '
'db1->db2->db3->db4-> '
'db5->db6->db7->db8-> '
'db9->db10->db**-> '
'=================================== '
'Symlets ->->sym '
'------------------------------ '
'sym2->sym3->sym4->sym5-> '
'sym6->sym7->sym8->sym**-> '
'=================================== '
'Coiflets ->->coif '
'------------------------------ '
'coif1->coif2->coif3->coif4-> '
'coif5-> '
'=================================== '
'BiorSplines ->->bior '
'------------------------------ '
'bior1.1->bior1.3->bior1.5->bior2.2-> '
'bior2.4->bior2.6->bior2.8->bior3.1-> '
'bior3.3->bior3.5->bior3.7->bior3.9-> '
'bior4.4->bior5.5->bior6.8-> '
'=================================== '
'ReverseBior ->->rbio '
'------------------------------ '
'rbio1.1->rbio1.3->rbio1.5->rbio2.2-> '
'rbio2.4->rbio2.6->rbio2.8->rbio3.1-> '
'rbio3.3->rbio3.5->rbio3.7->rbio3.9-> '
'rbio4.4->rbio5.5->rbio6.8-> '
'=================================== '
'Meyer ->->meyr '
'=================================== '
'DMeyer ->->dmey '
'=================================== '
'Gaussian ->->gaus '
'------------------------------ '
'gaus1->gaus2->gaus3->gaus4-> '
'gaus5->gaus6->gaus7->gaus8-> '
'=================================== '
'Mexican_hat ->->mexh '
'=================================== '
'Morlet ->->morl '
'=================================== '
'Complex Gaussian ->->cgau '
'------------------------------ '
'cgau1->cgau2->cgau3->cgau4-> '
'cgau5->cgau6->cgau7->cgau8-> '
'=================================== '
'Shannon ->->shan '
'------------------------------ '
'shan1-1.5->shan1-1->shan1-0.5->shan1-0.1-> '
'shan2-3->shan**-> '
'=================================== '
'Frequency B-Spline ->->fbsp '
'------------------------------ '
'fbsp1-1-1.5->fbsp1-1-1->fbsp1-1-0.5->fbsp2-1-1->'
'fbsp2-1-0.5->fbsp2-1-0.1->fbsp**-> '
'=================================== '
'Complex Morlet ->->cmor '
'------------------------------ '
'cmor1-1.5->cmor1-1->cmor1-0.5->cmor1-1-> '
'cmor1-0.5->cmor1-0.1->cmor**-> '
'=================================== '
'Fejer-Korovkin ->->fk '
'------------------------------ '
'fk4->fk6->fk8->fk14-> '
'fk18->fk22-> '
'=================================== '
'Best-localized Daubechies->->bl '
'------------------------------ '
'bl7->bl9->bl10-> '
'=================================== '
'Morris minimum-bandwidth ->->mb '
'------------------------------ '
'mb4.2->mb8.2->mb8.3->mb8.4-> '
'mb10.3->mb12.3->mb14.3->mb16.3-> '
'mb18.3->mb24.3->mb32.3-> '
'=================================== '
'Beylkin ->->beyl '
'=================================== '
'Vaidyanathan ->->vaid '
'=================================== '
'Han linear-phase moments ->->han '
'------------------------------ '
'han2.3->han3.3->han4.5->han5.5-> '
'=================================== '
'Lemarie ->->lem '
'------------------------------ '
'lem1->lem2->lem3->lem4-> '
'lem5-> '
'=================================== '
Restore the initial wavelets. Restore the initial ASCII file wavelets.ini and the initial MAT-file wavelets.bin. Regenerate the list of wavelet families. The list does not include the new family.
wavemngr('restore',0) wavemngr('read')
ans = 23x35 char array
'==================================='
'Haar ->->haar '
'Daubechies ->->db '
'Symlets ->->sym '
'Coiflets ->->coif '
'BiorSplines ->->bior '
'ReverseBior ->->rbio '
'Meyer ->->meyr '
'DMeyer ->->dmey '
'Gaussian ->->gaus '
'Mexican_hat ->->mexh '
'Morlet ->->morl '
'Complex Gaussian ->->cgau '
'Shannon ->->shan '
'Frequency B-Spline ->->fbsp '
'Complex Morlet ->->cmor '
'Fejer-Korovkin ->->fk '
'Best-localized Daubechies->->bl '
'Morris minimum-bandwidth ->->mb '
'Beylkin ->->beyl '
'Vaidyanathan ->->vaid '
'Han linear-phase moments ->->han '
'==================================='
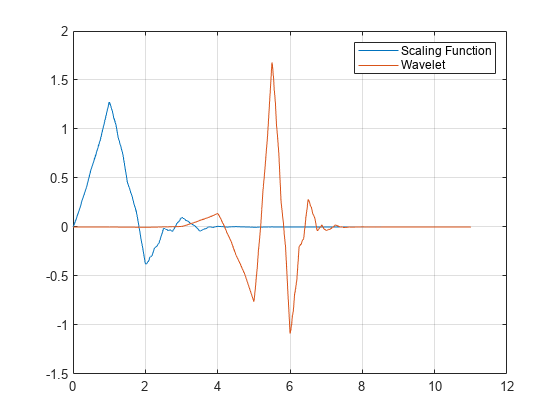
All command line capabilities are available for new families of wavelets. Create a new family. Compute the four associated filters and the scale and wavelet functions.
wavemngr('add','Lemarie','lem',1,'1 2 3 4 5','lemwavf'); [Lo_D,Hi_D,Lo_R,Hi_R] = wfilters('lem3'); [phi,psi,xval] = wavefun('lem3'); plot(xval,[phi;psi]); legend('Scaling Function','Wavelet') grid on
Remove the added family.
wavemngr('del','Lemarie')
Input Arguments
FN — Wavelet family name
character vector | string scalar
Wavelet family name, specified as a character vector or string scalar.
FSN — Wavelet family short name
character vector | string scalar
Wavelet family short name, specified as a character vector or string
scalar. The number of characters in FSN must be less
than or equal to 4.
WT — Wavelet family type
1 | 2 | 3 | 4 | 5
Wavelet family type, specified as one of the following:
1— Orthogonal wavelets2— Biorthogonal wavelets3— Wavelet with a scaling function4— Wavelet without a scaling function5— Complex wavelet without a scaling function
NUMS — Wavelet parameters
'' | character vector | string scalar
Wavelet parameters, specified as:
If the family consists of a single wavelet,
NUMSis the empty string''. For example, themexhandmorlfamilies each contain a single wavelet.If the wavelet is member of a finite family of wavelets,
NUMScontains a space-separated list of items representing wavelet parameters. For example, for the biorthogonal wavelet familybior,NUMS = '1.1 1.3 1.5 2.2 2.4 2.6 2.8 3.1 3.3 3.5 3.7 3.9 4.4 5.5 6.8'.If the wavelet is member of an infinite family of wavelets,
NUMScontains a space-separated list of items representing wavelet parameters, terminated by the special sequence**. Two examples are listed in the following table.Wavelet Family NUMSdbNUMS = '1 2 3 4 5 6 7 8 9 10 **'shanNUMS = '1-1.5 1-1 1-0.5 1-0.1 2-3 **'
TYPNUMS — Wavelet parameter input format
'integer' (default) | 'real' | 'charactervector'
Wavelet parameter input format, specified as:
'integer'— Use this option when the parameter is an integer. For example, the Daubechies wavelet familydbuses an integer parameter.'real'— Use this option when the parameter is real. For example, the biorthogonal wavelet familybioruses a real parameter.'charactervector'— Use this option when the parameter is a character vector. For example, the Shannon wavelet family uses a character vector.
FILE — Wavelet definition file
character vector | string scalar
Wavelet definition file, specified as a character vector or string scalar.
FILE is the name of a MAT-file or a code file name
that defines the wavelet family.
If the wavelet family contains only one type 1 (orthogonal) or type 2 (biorthogonal) wavelet, you can define the wavelet in a MAT-file. The MAT-file contains the scaling filter coefficients. The filename must match the wavelet family short name.
If you define an orthogonal wavelet in a MAT-file, the name of the variable containing the scaling filter coefficients must match the name of the wavelet family short name.
If you define a biorthogonal wavelet in a MAT-file, the names of the variables containing the scaling filter coefficients must be
DfandRf.
For more information, see Add Quadrature Mirror and Biorthogonal Wavelet Filters.
Example: If a family that contains a single orthogonal wavelet has the
short name wfsn, the variable wfsn
must contain the scaling filter coefficients. To create the necessary
MAT-file, you would use the command save wfsn
wfsn.
B — Effective support
two-element real-valued vector
Effective support for wavelets with family type WT
equal to 3, 4, or 5, specified as a two-element real-valued vector. If
B = [lb ub], then lb specifies the
lower bound, and ub specifies the upper bound.
Data Types: double
N — Wavelet
character vector | string scalar
Wavelet or wavelet family to delete, specified by a character vector or
string scalar. N is either the wavelet name or wavelet
family short name.
Example: wavemngr('del','Lemarie')
wname — Wavelet
character vector | string scalar
Wavelet, specified as a character vector or string scalar. To learn what
wavelets are available in a wavelet family, use waveinfo and the family
short name.
Example: ty = wavemngr('type',"coif4")
Output Arguments
out — Wavelet manager output
character array
Wavelet manager output, returned as a character array.
wavetype — Wavelet family type
integer
Wavelet family type, returned as an integer.
1— Orthogonal wavelets2— Biorthogonal wavelets3— Wavelet with a scaling function4— Wavelet without a scaling function5— Complex wavelet without a scaling function
Limitations
wavemngrallows you to add a wavelet. You must verify that it is truly a wavelet. No check is performed to confirm the addition is a wavelet or to confirm the type of the new wavelet. You can useisorthwfbandisbiorthwfbto determine if a wavelet is orthogonal or biorthogonal.
References
[1] Daubechies, I. Ten Lectures on Wavelets. CBMS-NSF Regional Conference Series in Applied Mathematics. Philadelphia, PA: Society for Industrial and Applied Mathematics, 1992.
Version History
Introduced before R2006a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)