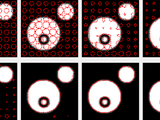
Active Contour Toolbox
This toolbox provides some functions for manipulating planar, closed splines to implement image or video segmentation by means of deformable (or active) contours. Contour topology is managed in a way that should allow changes similar to what can be observed with level sets (merging and splitting but no hole creation). Several objects can be segmented simultaneously in several frames.
Cite As
Eric Debreuve (2024). Active Contour Toolbox (https://www.mathworks.com/matlabcentral/fileexchange/11643-active-contour-toolbox), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Image Processing and Computer Vision > Image Processing Toolbox > Image Segmentation and Analysis > Image Segmentation > Active contours >
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
acontour/
acontour/conversion/
acontour/instantiation/
acontour/interface/
acontour/interface/private/
acontour/plot/
acontour/property/dynamic/
acontour/property/static/
acontour/transformation/
acontour/usage/
polygon/
polygon/conversion/
polygon/instantiation/
polygon/plot/
polygon/property/
polygon/transformation/
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.0.0.0 |