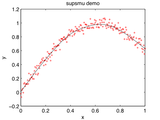
This is a MATLAB version of Jerome Friedman's 1984 supersmoother algorithm. The original was written in Fortran; this is a vectorized translation. The algorithm is a variable span smoother which uses cross validation to pick the best span for each predicted point.
Cite As
Douglas Schwarz (2025). Supersmoother (https://www.mathworks.com/matlabcentral/fileexchange/17986-supersmoother), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Created with
R2007b
Compatible with any release
Platform Compatibility
Windows macOS LinuxCategories
Find more on Smoothing in Help Center and MATLAB Answers
Tags
Acknowledgements
Inspired: ReSpect
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.0.0.0 |