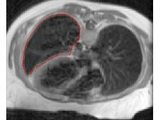
Snakes: Active Contour Models
This demo implements the Active Contour Models as proposed by Kass et al.
To run it with GUI
1. Type guide on the matlab prompt.
2. Click on "Go to Existing GUI"
3. Select the snk.fig file in the same directory as this file
4. Click the green arrow at the top to launch the GUI
Once the GUI has been launched, you can use snakes by
1. Click on "New Image" and load an input image. Samples image are provided.
2. Set the smoothing parameter "sigma" or leave it at its default value and click "Filter". This will smooth the image.
3. As soon as you click "Filter", cross hairs would appear and using them and left click of you mouse you can pick initial contour location on the image. A red circle would appear everywhere you click and in most cases you should click all the way around the object you want to segment. The last point must be picked using a right-click in order to stop matlab for asking for more points.
4. Set the various snake parameters (relative weights of energy terms in the snake objective function) or leave them with their default value and click "Iterate" button. The snake would appear and move as it converges to its low energy state.
Copyright (c) Ritwik Kumar, Harvard University 2010
www.seas.harvard.edu/~rkkumar
This code implements “Snakes: Active Contour Models” by Kass, Witkin and Terzopolous incorporating Eline, Eedge and Eterm energy factors. See the included report and the paper to learn more.
If you find this useful, also look at Radon-Like Features based segmentation in the following paper:
Ritwik Kumar, Amelio V. Reina & Hanspeter Pfister, Radon-Like Features and their Application to Connectomics”, IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA) 2010
http://seas.harvard.edu/~rkkumar
Its code is also available on MATLAB Central
Cite As
Ritwik Kumar (2024). Snakes: Active Contour Models (https://www.mathworks.com/matlabcentral/fileexchange/28109-snakes-active-contour-models), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Simulink > Modeling >
- Sciences > Biological and Health Sciences > Biomedical Imaging >
- Image Processing and Computer Vision > Image Processing Toolbox > Image Segmentation and Analysis > Image Segmentation > Active contours >
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
activeContoursDemo/
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.0.0.0 |