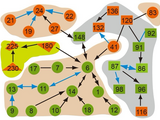
Ancestral polytree
Causal polytrees are singly connected causal models and they are
frequently applied in practice. However, in various applications, many
variables remain unobserved and causal polytrees cannot be applied
without explicitly including unobserved variables. Our study thus
proposes the ancestral polytree model, a novel combination of
ancestral graphs and singly connected graphs. Ancestral graphs can
model causal and non-causal dependencies, while singly connected
models allow for efficient learning and inference. We discuss the
basic properties of ancestral polytrees and propose an efficient
structure learning algorithm. Experiments on synthetic datasets and
biological datasets show that our algorithm is efficient and the
applications of ancestral polytrees are promising.
Citation: Guangdi Li, Anne-Mieke Vandamme, Jan Ramon, Learning Ancestral Polytrees. The workshop of Learning Tractable Probabilistic Model co-located with the 31st International Conference on Machine Learning (ICML 2014), Beijing, China, 2014 https://lirias.kuleuven.be/handle/123456789/456333
Cite As
Guangdi Li (2024). Ancestral polytree (https://www.mathworks.com/matlabcentral/fileexchange/40126-ancestral-polytree), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.