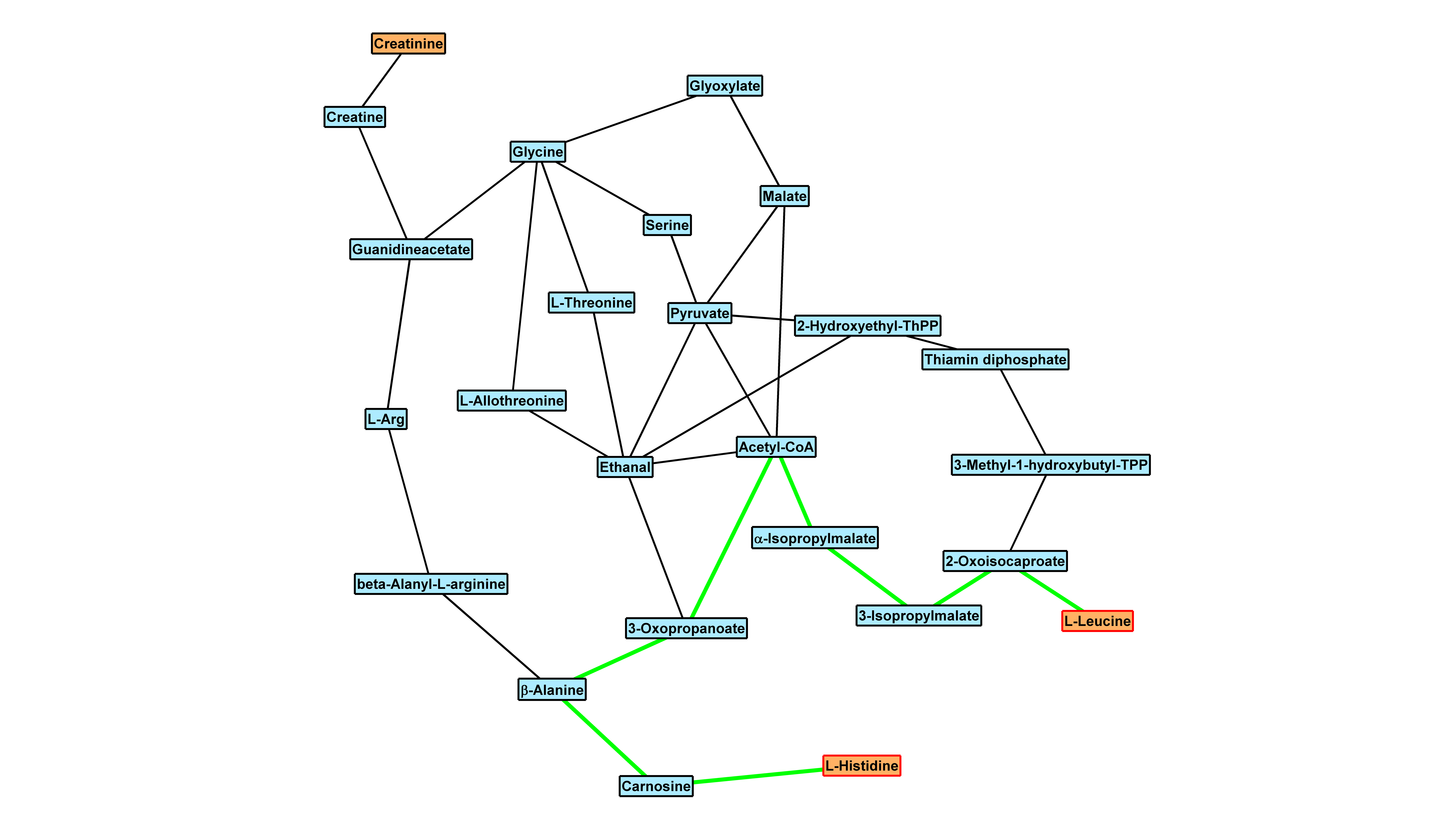
MetaboNetworks
A Matlab-based tool to create custom sub-networks using main substrate-product pairs as defined by the Kyoto Encyclopaedia of Genes and Genomes (KEGG). It can be used to explore transgenomic interactions, for example mammalian and bacterial associations. It calculates the shortest path between a set of metabolites (e.g. biomarkers from a metabonomic study) and plots the connectivity between metabolites as links in a network graph. The resulting graph can be changed and explored interactively. Furthermore, nodes in the graph are linked to the KEGG compound webpage.
A multi-systemic example data set is provided of a supra-organism, this includes reactions that can occur in compartments of homo sapiens, firmicutes and/or bacretoidetes species. The MetaboGetworks function can be used to generate custom multi-organism databases to use in the MetaboNetworks function. (See the READ ME file)
Cite As
Joram Posma (2024). MetaboNetworks (https://www.mathworks.com/matlabcentral/fileexchange/42684-metabonetworks), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
Tags
Acknowledgements
Inspired by: MatlabBGL
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 2.1.0.0 | Updated functions GetRPData and GetRPData_Outer to reflect changes in KEGG. Added a new example data set. |
||
| 1.4.0.0 | Fixed the error in OrganismFindGUI caused by a naming ambiguity in KEGG.
|
||
| 1.3.0.0 | 2 new functionalities: edges linked to corresponding KEGG reaction pair web page, text edit option to change a specific string (e.g. 'hydroxy' to 'OH') for all nodes simultaneously. Search functions for selecting organisms/metabolites were updated. |
||
| 1.2.0.0 | Fixed 2 errors in subfunctions of MetaboGetworks, added function descriptions of input and output variables. |
||
| 1.1.0.0 | Removal of a parfor in the TextMatch code. |
||
| 1.0.0.0 |