Tools for NIfTI and ANALYZE image
Editor's Note: As of Release 2017b, MATLAB now provides support for reading and writing NIfTI image data files in the Image Processing Toolbox
Please always check NIfTI_tools.pdf for detail descriptions and latest updates.
If you are confused by the Left / Right of an ANALYZE image, please read UseANALYZE.pdf.
You may also want to go through FAQ.pdf for practical solutions and real examples.
Basic Programs:
1. load_untouch_header_only.m: Load only the header section of NIfTI or ANALYZE file. The input file will be automatically detected. NIfTI structure will be returned for NIfTI file, and ANALYZE structure will be returned for ANALYZE file.
2. load_nii.m: Load N-Dimensional NIfTI file (where N can be from 3 to 7) or ANALYZE file (where N can be from 3 to 4), and apply header info (e.g. affine geometric transform, voxel intensity scaling, etc.) to the data. If your file has more than 3-Dimension (e.g. time series etc.), you can also specify a range to extract only 1 or several volumes.
3. save_nii.m: Save N-Dimensional NIfTI structure (where N can be from 3 to 7) that is loaded by "load_nii.m" or made by "make_nii.m" into a NIfTI file.
4. make_nii.m: Make N-Dimensional NIfTI structure (where N can be from 3 to 7) based on the N-Dimensional matrix and other optional parameters (e.g. voxel_size, origin, etc.). Using "save_nii" command, the NIfTI structure that is made by "make_nii" can be saved into a NIfTI file.
5. make_ana.m: Make 3D ANALYZE structure based on the 3D matrix and other optional parameters (e.g. voxel_size, origin, etc.). Using "save_untouch_nii" command, the ANALYZE structure that is made by "make_ana" can be saved into an ANALYZE file in order to be compatible with some ANALYZE only programs.
6. reslice_nii.m: Re-sample 3D (or 4D) NIfTI file, or ANALYZE file with affine matrix M in .mat file, and save the re-sampled data into a new NIfTI file. The program will base on the affine matrix, which is especially useful for oblique images with non-orthogonal rotation or shearing that cannot be loaded with "load_nii.m". You can also specify voxel_size, etc. It will not cause negative effect, as long as you remember not to do slice time correction after using "reslice_nii.m".
7. pad_nii.m: Pad the volume(s) in NIfTI structure from any of the six sides, while keeping the originator, voxel size, data type, and description unchanged. The program is especially useful after you use reslice_nii, since the new volume will most likely have different dimensions.
8. clip_nii.m: Clip the volume(s) in NIfTI structure from any of the six sides, while keeping the originator, voxel size, data type, and description unchanged. The program is especially useful after you use reslice_nii, since the new volume will most likely have different dimensions.
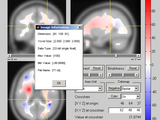
9. view_nii.m: View & Edit 3D (or 4D) NIfTI or ANALYZE structure that is loaded by "load_nii.m" or made by "make_nii.m". Activation map, ROI, etc. can be overlaid on top of a background image (see above picture). Plotted view can be embedded into your existing figure window. If you use it as an individual program, it can also edit the orientation and voxel value of the image, view volume histogram, and save the modified image.
10. load_untouch_nii.m: Load N-Dimensional NIfTI file (where N can be from 3 to 7) or ANALYZE file (where N can be from 3 to 4), but do not apply any changes that are indicated in the header. WARNING: Do not use "view_nii.m" to view the structure that is loaded by "load_untouch_nii.m".
11. save_untouch_nii.m: Save N-Dimensional NIfTI structure (where N can be from 3 to 7) or ANALYZE structure (where N can be from 3 to 4) that is loaded by "load_untouch_nii.m" or made by "make_ana.m" into a new NIfTI or ANALYZE file. If you do not modify the loaded dataset, the header and data in the new saved file should be the same as those in the original file.
Other Programs:
1. collapse_nii_scan.m: Integrate multiple single-scan NIfTI or ANALYZE files into a multiple-scan NIfTI file.
2. expand_nii_scan.m: Break a multiple-scan NIfTI file into multiple single-scan NIfTI files.
3. save_untouch_slice.m: Save back to the original image with a portion of slices that was loaded by load_untouch_nii. You can process those slices matrix in any way, as long as their dimension is not altered.
4. get_nii_frame.m: Return the number of time frames of a NIfTI file.
5. flip_lr.m: Flip NIfTI or ANALYZE file Left-Right along the plane across the originator, and save the L-R flipped data into a NIfTI file. WARNING: Please use this program with caution, although you can always flip it back.
6. load_nii_ext.m: Load header extension from a NIfTI file.
7. mat_into_hdr.m: Integrate affine matrix in old SPM .mat file into its .hdr header file. Thus, the ANALYZE file is converted into a NIfTI file with the updated .hdr header file.
Cite As
Jimmy Shen (2024). Tools for NIfTI and ANALYZE image (https://www.mathworks.com/matlabcentral/fileexchange/8797-tools-for-nifti-and-analyze-image), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Image Processing and Computer Vision > Image Processing Toolbox > Import, Export, and Conversion >
- Sciences > Neuroscience > Human Brain Mapping > MRI >
- Image Processing and Computer Vision > Image Processing Toolbox > Get Started with Image Processing Toolbox >
- Industries > Medical Devices > DICOM Format >
Tags
Acknowledgements
Inspired by: MRI Analyze tools
Inspired: PCG_unwrap_2D(ph_nii, ph_file, mask_nii, max_iter, epsi_con, N), MRIqual, BioImage Suite Web, ExportPngInkscape(filename,dpi,handle), Parametric Mapping Scripts for MRI data, fMRIData Tool, NIfTI Studio, Visualization of multi-modal volume segmentation, Siemens DICOM sort and convert to NIfTI, Bipolar Colormap, Generate synthetic fMRI data, RAPID: a Routine Assurance Pipeline for Imaging of Diffusion, 3D NIfTI Data Viewer, Seconds to hours, minutes, seconds, Composite Image, create_mosaic.m, Field Mapping Toolbox, Function for Viewing MRI Slices, A GUI to load and view .NII files., DICOM to NIfTI converter, Finding functional networks in brain fMRI data using stepwise clustering, Brain Ruler
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.27.0.0 | 1. Thanks to the feedback and contribution of Chris McNorgan from Western University, the rri_orient.m now have optional parameter for orientation vector.
|
||
| 1.26.0.0 | Since voxel size can be less than 0, I will change the default voxel_size in reslice_nii.m from rounded minimum voxel_size in original NIfTI header to absolute minimum voxel_size in original NIfTI header. This fixed reslice_nii bug. |
||
| 1.25.0.0 | Added some new features and fixed some bugs. For more detail, please visit: |
||
| 1.24.0.0 | Thanks to the feedback of Matthew Bickell from University of Cape Town, a bug is fixed when edit voxel in millimeter unit. |
||
| 1.23.0.0 | "make_nii.m" is now able to automatically detect both single and double complex data type. |
||
| 1.22.0.0 | Fixed a bug for yesterday's release |
||
| 1.21.0.0 | You can now create RGB NIfTI / ANALYZE file structure using make_nii / make_ana program. |
||
| 1.20.0.0 | New program "load_untouch_header_only.m" and "save_untouch_slice.m" is provided. Check http://www.rotman-baycrest.on.ca/~jimmy/NIFTI for more details. |
||
| 1.19.0.0 | New feature is added in "view_nii" program to let you edit voxel values or set landmarks. Also thanks to the code from Roman, which speed up "load_untouch_nii_img" program when the slices are not loaded at once. |
||
| 1.16.0.0 | 1. Fixed an overlay problem in view_nii.m, so the color of min/max is now showing properly.
|
||
| 1.14.0.0 | Thanks to the feedback of Roman Fleysher from Yeshiva University, I fixed a bug when loading a quaternion form NIfTI data. |
||
| 1.13.0.0 | Thanks to Rembrandt from Radboud University Nijmegen (Netherlands), I made 2 small changes to handle the exceptions. |
||
| 1.12.0.0 | Fixed a bug in "collapse_nii_scan" to collapse Analyze format data. |
||
| 1.11.0.0 | 1. "load_untouch_nii" now supports loading specific slices of NIfTI or Analyze format.
|
||
| 1.10.0.0 | "load_untouch_nii" now supports loading specific slices of NIfTI or Analyze format. |
||
| 1.9.0.0 | While an N-Dimensional matrix can be saved into a NIfTI file using "make_nii / save_nii" command pair, a 3D matrix can also be saved into an ANALYZE file using "make_ana / save_untouch_nii" in order to be compatible with some ANALYZE only programs. |
||
| 1.8.0.0 | Fixed several bugs, and posted informative table: |
||
| 1.7.0.0 | Just receive warning from MATLAB: "If I do not license my code with BSD, my code will be removed." |
||
| 1.6.0.0 | Thanks to the feedback of Kate Fissell from University of Pittsburgh, transformation matrix M will no longer be saved by save_untouch_nii.m. |
||
| 1.5.0.0 | 1. Thanks to the feedback of Bryce Wilkins from USC, a typo in one of my message display is fixed. 2. Thanks to the suggestion of Ashish Raj from Cornell, 'expand_nii_scan.m' is modified and 'collapse_nii_scan.m' is added. |
||
| 1.4.0.0 | Default data type for 'make_nii.m' now follows the data type of 'img' matrix, instead of float32. |
||
| 1.3.0.0 | Fixed a bug in fliplr.m when reading files. |
||
| 1.2.0.0 | update didn't show up |
||
| 1.1.0.0 | 1. Properly handle fread/'*char' problem with foreign characters in MATLAB 7 and above; 2. Reordered input parameters for 'reslice_nii.m' program; |
||
| 1.0.0.0 | Extended Load / Save / Make programs to N-Dimension, where N can be from 3 to 7 for NIfTI data and from 3 to 4 for ANALYZE data. |