resubPredict
Predict response of regression tree by resubstitution
Syntax
Description
Examples
Compute the In-Sample MSE
Load the carsmall data set. Consider Displacement, Horsepower, and Weight as predictors of the response MPG.
load carsmall
X = [Displacement Horsepower Weight];Grow a regression tree using all observations.
Mdl = fitrtree(X,MPG);
Compute the resubstitution MSE.
Yfit = resubPredict(Mdl); mean((Yfit - Mdl.Y).^2)
ans = 4.8952
You can get the same result using resubLoss.
resubLoss(Mdl)
ans = 4.8952
Estimate In-Sample Responses for Each Subtree
Load the carsmall data set. Consider Weight as a predictor of the response MPG.
load carsmall
idxNaN = isnan(MPG + Weight);
X = Weight(~idxNaN);
Y = MPG(~idxNaN);
n = numel(X);Grow a regression tree using all observations.
Mdl = fitrtree(X,Y);
Compute resubstitution fitted values for the subtrees at several pruning levels.
m = max(Mdl.PruneList);
pruneLevels = 1:4:m; % Pruning levels to consider
z = numel(pruneLevels);
Yfit = resubPredict(Mdl,Subtrees=pruneLevels);Yfit is an n-by-z matrix of fitted values in which the rows correspond to observations and the columns correspond to a subtree.
Plot several columns of Yfit and Y against X.
sortDat = sortrows([X Y Yfit],1); % Sort all data with respect to X plot(repmat(sortDat(:,1),1,size(Yfit,2)+1),sortDat(:,2:end)) % Vectorize for efficiency lev = num2str((pruneLevels)',"Level %d MPG"); legend(["Observed MPG"; lev]) title("In-Sample Fitted Responses") xlabel("Weight (lbs)") ylabel("MPG") h = findobj(gcf); set(h(4:end),LineWidth=3) % Widen all lines
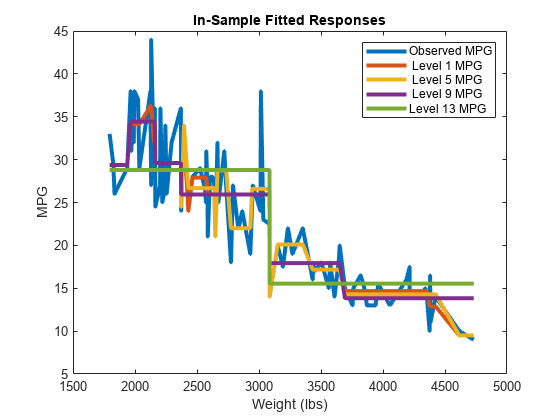
The values of Yfit for lower pruning levels tend to follow the data more closely than higher levels. Higher pruning levels tend to be flat for large X intervals.
Input Arguments
tree — Regression tree model
RegressionTree model object
Regression tree model, specified as a RegressionTree model object trained with fitrtree.
subtrees — Pruning level
0 (default) | vector of nonnegative integers | "all"
Pruning level, specified as a vector of nonnegative integers in ascending
order or "all".
If you specify a vector, then all elements must be at least
0 and at most max(tree.PruneList).
0 indicates the full, unpruned tree, and
max(tree.PruneList) indicates the completely pruned
tree (in other words, just the root node).
If you specify "all", then
resubPredict operates on all subtrees (in other
words, the entire pruning sequence). This specification is equivalent to
using 0:max(tree.PruneList).
resubPredict prunes tree to each
level specified by Subtrees, and then estimates the
corresponding output arguments. The size of subtrees
determines the size of some output arguments.
For the function to invoke subtrees, the properties
PruneList and PruneAlpha of
tree must be nonempty. In other words, grow
tree by setting Prune="on" when
you use fitrtree, or by pruning
tree using prune.
Data Types: single | double | char | string
Output Arguments
Yfit — Predicted resubstitution response values
numeric vector | numeric matrix
Predicted resubstitution response values for the training data, returned
as a numeric vector or a numeric matrix. Yfit is of the
same data type as the training response data
tree.Y.
If Subtrees is a numeric scalar, then
Yfit is returned as a numeric column vector.
Otherwise, Yfit is returned as a matrix with
m columns, where m is the number
of subtrees. Each column represents the predictions of the corresponding
subtree.
node — Node numbers
numeric column vector | numeric matrix
Node numbers for the predicted classes, returned as a numeric column vector or numeric matrix.
If subtrees is a scalar or is not specified, then
resubPredict returns node as a numeric column
vector with n rows, the same number of rows as
tree.X.
If subtrees contains m > 1
entries, then node is an n-by-m
numeric matrix. Each column represents the node predictions of the corresponding
subtree.
Extended Capabilities
GPU Arrays
Accelerate code by running on a graphics processing unit (GPU) using Parallel Computing Toolbox™.
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2011a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.

Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list:
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)