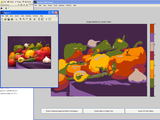
clusterImg
[IDX,C,sumd,D] = clusterImg(imgin,nClusters,inMask)
This function implements kmeans clustering on an input RGB (m x n x 3) image. The user inputs at least two inputs: IMGIN and NCLUSTERS, and this function will step through an interactive color segmentation using kmeans clustering. It is interactive in that the user will be prompted to click-define the target colors in the segmentation. (There will be NCLUSTERS prompts.) Optionally, the user can also input a third argument containing a binary mask of the input image, in which case the program will operate on a subimage defined by the BOUNDING BOX of the (true) inMask.
Following segmentation (clustering), the user can EXPORT THE CLUSTERED IMAGE TO THE WORKSPACE, along with its corresponding colormap. Additionally, the user can opt to EXPORT A BINARY MASK OF A SINGLE COLOR CLUSTER TO THE WORKSPACE, or AUTO-GENERATE ALL MASKS. All of these options are initiated by press of a uicontrol pushbutton.
Cite As
Brett Shoelson (2026). clusterImg (https://www.mathworks.com/matlabcentral/fileexchange/12979-clusterimg), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Image Processing and Computer Vision > Image Processing Toolbox > Image Segmentation and Analysis > Image Segmentation >
Tags
Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.3.0.1 | Updated license |
||
| 1.3.0.0 | Updated M file and license text file copyright to The MathWorks, Inc. |
||
| 1.2.0.0 | updated license text file to The MathWorks, Inc. |
||
| 1.1.0.0 | added copyright |
||
| 1.0.0.0 | New pushbutton will auto-generate binary masks for all (clustered) colors in the image. Also, fixes two small bugs (missing documentation for LABELS, improper capitalization in description of workspace variables.) |