fexact( varargin )
Matlab now (2014b) has a two-tailed Fisher exact test (fishertest)! However, I still need a version that can be vectorized to perform 1000s of tests. This implementation performs 2.5e6 one or two-tailed tests per second and is ideally suited for genome-wide association studies or other large problems where 100s of samples and up to 100s of 1000s of tests are being performed.
It has been verified correct for samples sizes up to 100 and spot checked beyond that up to 1000s of samples.
Cite As
Michael Boedigheimer (2024). fexact( varargin ) (https://www.mathworks.com/matlabcentral/fileexchange/22550-fexact-varargin), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.12.0.0 | Improved speed for large number of observations.
|
||
| 1.11.0.0 | Improved speed for large number of observations.
|
||
| 1.10.0.0 | Chris Rorden added support for single-sided Leibermeister tests, which corrects for uncertaintity in the marginal sums.
|
||
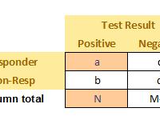
| 1.8.0.0 | changed to allow permutation testing in 2012a. Added a graphic to help explain contingency table |
||
| 1.6.0.0 | fixed bug in detecting error conditions |
||
| 1.5.0.0 | Fixed a reporting error that may affect results when a single test is done. The error that allowed p-values to be larger than 1 to be reported. p-values smaller than one should not be affected. |
||
| 1.4.0.0 | Has been updated to work with a large sample size if performing a single test. |
||
| 1.3.0.0 | Now can handle large number of observations in a single trial with minimal footprint.
|
||
| 1.2.0.0 | Improved documentation and help. Improved performance for single tests. |
||
| 1.0.0.0 |