Feature Selection using Matlab
The DEMO includes 5 feature selection algorithms:
• Sequential Forward Selection (SFS)
• Sequential Floating Forward Selection (SFFS)
• Sequential Backward Selection (SBS)
• Sequential Floating Backward Selection (SFBS)
• ReliefF
Two CCR estimation methods:
• Cross-validation
• Resubstitution
After selecting the best feature subset, the classifier obtained can be used for classifying any pattern.
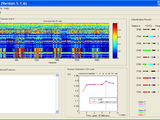
Figure: Upper panel is the pattern x feature matrix
Lower panel left are the features selected
Lower panel right is the CCR curve during feature selection steps
Right panel is the classification results of some patterns.
This software was developed using Matlab 7.5 and Windows XP.
Copyright: D. Ververidis and C.Kotropoulos
AIIA Lab, Thessaloniki, Greece,
jimver@aiia.csd.auth.gr
costas@aiia.csd.auth.gr
In order to run the DEMO:
In order to run the demo:
- A PC with Windows XP is needed.
- Use Matlab7.5 or later to run DEMO.m
1) Select the ‘finalvec.mat’ dataset (patterns x [features+1] matrix) from 'PatTargMatrices' folder. The last column of ‘finalvec.mat’ are the targets.
2) Press the run button on the panel. It is the second one.
3) After the selection of the optimum feature set, select a set of patterns for classification using the open folder button (last button). It can be the same data-set that was used for training the feature selection algorithm
% REFERENCES:
[1] D. Ververidis and C. Kotropoulos, "Fast and accurate feature subset selection applied into speech emotion recognition," Els. Signal Process., vol. 88, issue 12, pp. 2956-2970, 2008.
[2] D. Ververidis and C. Kotropoulos, "Information loss of the Mahalanobis distance in high dimensions: Application to feature selection," IEEE Trans. Pattern Analysis and Machine Intelligence, vol. 31, no. 12, pp. 2275-2281, 2009.
Cite As
Dimitrios Ververidis (2026). Feature Selection using Matlab (https://www.mathworks.com/matlabcentral/fileexchange/22970-feature-selection-using-matlab), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
Tags
Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
Version_5.1.8_Out/
Version_5.1.8_Out/LowLevelFunctions/
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.5.0.0 | PAMI paper accepted |
||
| 1.4.0.0 | 5.1.8 Help menu added
|
||
| 1.3.0.0 | News: Version 5.1.6 supports up to 7 classes. |
||
| 1.1.0.0 | Latest version 5.1.5 includes also
|
||
| 1.0.0.0 |