Developing in vivo Functional Imaging Technology with Micron-Scale Resolution Using Optical Coherence Tomography
By Orly Liba, Elliott D. SoRelle, and Adam de la Zerda, Stanford University
Researchers and physicians rely on functional imaging to better understand tumors and other structures within the human body. However, imaging technologies that capture deep structures have poor resolution, while those that provide high resolution have limited depth. Positron emission tomography (PET), for example, reveals details deep within tissue but suffers from poor spatial resolution, with each voxel of a PET scan representing thousands or even millions of cells. In contrast, optical microscopy can deliver subcellular spatial resolution but is usually limited to a depth of tens of microns.
Optical coherence tomography (OCT) helps bridge the gap between low-resolution/high-penetration and high-resolution/low-penetration technology by providing micron-scale spatial resolution at depths of one to two millimeters. Traditional OCT provides only structural information; it lacks the necessary contrast to provide functional or molecular information.
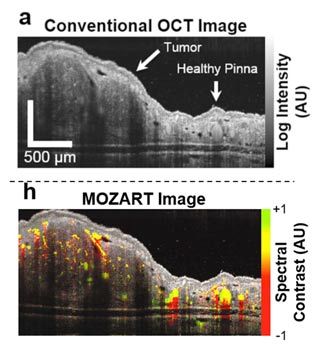
Our research group at Stanford has addressed this drawback by developing MOZART (MOlecular imaging and characteriZation of tissue noninvasively At cellular ResoluTion), a method that uses large gold nanorods (LGNRs) to improve the contrast of OCT images in vivo (Figure 1).
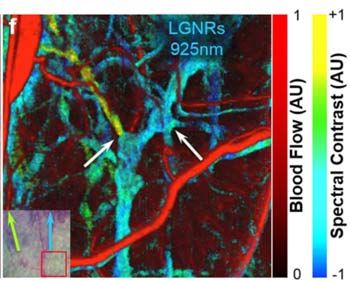
Using spectral processing algorithms developed in MATLAB®, we have analyzed the backscattering from these LGNRs to noninvasively image blood vessels as small as 20 μm in diameter and up to 750 μm deep in tumor tissue. The MATLAB algorithms in MOZART adaptively correct for dispersion and depth-related aberrations in every image, enabling us to identify individual capillaries as well as the locations and functional states of the valves that control fluid flow in lymphatic vessel networks. This information could help scientists detect and develop treatments for certain forms of cancer and blindness.
We could have developed our algorithms using Python® or another scripting language, but we chose MATLAB because all the functions and capabilities we need―basic matrix operations, image processing, signal processing, and more―are readily available. Furthermore, MATLAB includes convenient debugging capabilities that allow deeper exploration of our analysis algorithms.
Our MATLAB code is available for download.
Setting up the Experiments and Collecting Data
We demonstrated the capabilities of MOZART in two types of experiments in which we imaged the ears of living mice. For the first type of experiment, we injected LGNRs intravenously and imaged blood vessels in tumors and healthy tissue before and after the injection. In healthy subjects, the LGNRs circulate until they are processed by the liver and spleen. In subjects with tumors, the LGNRs tend to accumulate in the tumor due to the enhanced permeability and retention (EPR) effect.
For the second type of experiment, we injected the LGNRs subcutaneously (below the skin) and imaged their clearance into the lymph vessels. To study the performance of lymphatic valves, we sequentially injected two distinct types of LGNRs and then tracked each type as it passed through the lymphatic system. We were able to distinguish between the two types of LGNRs owing to their distinct scattering spectra and to our spectral algorithms, which were able to differentiate between them.
In both types of experiments, we used a broad superluminescent diode (SLD) to illuminate the tissue and a spectrometer to measure the backscattered light from the tissue and LGNRs, which are approximately 100 nm × 30 nm. The spectrometer records an interferogram, capturing the near-infrared scattering spectra from each point in the sample.
Developing Spectral Processing Algorithms
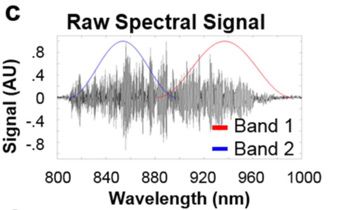
Once we had captured the raw interferogram data from the in vivo OCT scans, we developed algorithms to automate data processing. The algorithms reconstruct a conventional OCT image (like the one shown in Figure 1) from the recorded interferogram. They apply a discrete Fourier transform, implemented with matrix multiplication, to map the sample’s scatterers, which include both LGNRs and organic tissue in the sample.
Next, we used the unique scattering spectrum of the LGNRs to distinguish the LGNRs from the surrounding tissue. We updated the algorithms, applying Hann filters to divide the recorded spectrum into two bands (Figure 3).
After reconstructing images from these two bands and applying a median filter from Image Processing Toolbox™ to reduce noise, the algorithms compare the two images by performing a straightforward subtraction. When no LGNRs are present, the images reconstructed from the two bands are virtually identical, and the result of this subtraction is close to zero. However, when LGNRs are present, the two images differ significantly because of the distinct spectral scattering properties of the LGNRs.
We found that our ability to evaluate the differences between the images with this simple subtraction was hampered by two physical phenomena. The first was optical dispersion, which can be caused by optical elements in the OCT system and the sample itself. To compensate for dispersion, we added an iterative MATLAB algorithm that optimizes the alignment between the two reconstructed images for each sample we analyze.
The second issue we identified was due to depth-dependent spectral artifacts in the reconstructed difference image. These artifacts were primarily caused by chromatic aberrations introduced by the optical setup. To correct for this issue, we added an algorithm that measures the color gradient in a spectrally neutral region of the image and calculates a depth-dependent gain by fitting the gradient to a polynomial using the MATLAB polyfit function. This approach adaptively calibrates the depth-dependent spectral shift for each image.
After applying dispersion compensation and depth correction, the LGNRs showed up much more clearly because they significantly and consistently produced a higher spectral signal than the surrounding tissue.
Unexpected Results and Next-Generation Algorithms
The images produced by our MATLAB algorithms were so clear that they revealed a number of details that we did not fully anticipate seeing when we began our research. For example, we discovered instances in which LGNRs moved from blood vessels to lymph vessels. In a healthy subject such movement is unexpected, but this effect may have been due to porous blood vessels or an immune response. We were also surprised by how well we could see the lymph vessels, including the valves that control one-directional lymph flow in healthy subjects. To our knowledge, researchers have been unable to visualize lymph vessels and their functionality in this way until now.
We are currently enhancing our algorithms to support studies in which the LGNRs are coated with antibodies or peptides so that they target specific proteins in tumors. In our current studies, LGNRs are in motion as they flow through blood and lymph vessels. This flow makes it easy for our algorithms to average out noise. However, in studies where targeting is used, the LGNRs will remain localized in the tumor, making noise a more significant problem. We have already begun improving the noise-reduction capabilities of our next-generation algorithms in MATLAB to better visualize static LGNRs in preparation for future molecular targeting studies.
Acknowledgement
We gratefully recognize the work of our colleague Dr. Debasish Sen and his contribution to this research.
Published 2017 - 93073v00