incrementalPCA
Description
The incrementalPCA function creates an
incrementalPCA model object that is suitable for incremental principal
component analysis (PCA). Unlike the pca function, for which you must provide all of the data before computing the
principal component coefficients, incrementalPCA allows you to update the
coefficients incrementally by supplying chunks of data to the incremental fit
function.
Unlike other Statistics and Machine Learning Toolbox™ model objects, incrementalPCA can be called directly. Also,
you can specify incremental PCA options, such as the estimation period, warm-up period,
variable weights, and whether to standardize the predictor data before fitting the model to
data. After you create an incrementalPCA object, it is prepared for incremental
PCA.
Creation
You can create an incrementalPCA model object in two ways:
Call the function directly — Configure incremental PCA options by calling
incrementalPCAdirectly. This approach is best when you do not have data yet or you want to start incremental PCA immediately. When you callincrementalPCA, you can specify principal component coefficients and variances so that the initial model is warm.Call the incremental
fitfunction —fitaccepts a configuredincrementalPCAmodel object and data as input, and returns anincrementalPCAmodel object updated with information computed from the input model and data.
Description
IncrementalMdl = incrementalPCAIncrementalMdl. Properties of a
default model contain placeholders for unknown model parameters.
IncrementalMdl = incrementalPCA(Name=Value)incrementalPCA(StandardizeData=true,EstimationPeriod=1000) specifies
to standardize the predictor data using a hyperparameter estimation period of
1000 observations.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: IncrementalMdl =
incrementalPCA(StandardizeData=true,CenterData=true) specifies to standardize
and center the values of each variable.
Flag to center the data, specified as logical 0
(false) or 1 (true). If
CenterData=true, the incremental fit
function estimates the predictor means Mu during the estimation
period specified by EstimationPeriod and
subtracts them from the data before computing singular value decomposition or
eigenvalue decomposition.
If you specify CenterData=true, the number of degrees of
freedom is NumTrainingObservations – 1. Otherwise, the
number of degrees of freedom is NumTrainingObservations.
If you specify StandardizeData=true, the software sets
CenterData=true.
You cannot specify CenterData if you specify Coefficients and Latent.
Example: CenterData=true
Data Types: logical
Column means, specified as a numeric row vector.
If you specify Means:
You must also specify
Coefficients,Latent, andNumObservations.The
Muproperty ofIncrementalMdlis updated with the values inMeans.
Example: Means=[0 0 0.5 0.5 0.5]
Data Types: single | double
Number of observations, specified as a positive integer.
If you specify NumObservations, you must also specify
Coefficients and Latent.
If you also specify Means, the number of degrees of freedom
is NumObservations + NumTrainingObservations – 1. Otherwise, the
number of degrees of freedom is NumObservations +
NumTrainingObservations.
Example: NumObservations=100
Data Types: single | double
Flag to standardize the predictor data, specified as logical
0 (false) or 1
(true). If StandardizeData=true, the
incremental fit function estimates the predictor means
Mu and standard deviations Sigma during the
estimation period specified by EstimationPeriod, and
standardizes the predictor data.
If you specify StandardizeData=true, the number of degrees
of freedom is NumTrainingObservations - 1. Otherwise, the number
of degrees of freedom is NumTrainingObservations.
You cannot specify StandardizeData if you specify Coefficients and Latent.
Example: StandardizeData=true
Data Types: logical
Properties
You can set most properties by using name-value argument syntax when you call
incrementalPCA directly. You cannot set the properties
ExplainedVariance, IsWarm,
Mu, NumTrainingObservations, and
Sigma.
This property is read-only.
Principal component coefficients, specified as a
p-by-n numeric matrix, where
p is equal to NumPredictors and
n is equal to NumComponents. If you
specify Coefficients as a
p-by-n numeric matrix when creating the model
object, incrementalPCA sets NumPredictors=p and
NumComponents=n. Each column of Coefficients
contains coefficients for one principal component.
The incremental fit function updates
Coefficients and reorders the columns in descending order by the
principal component variances (see Latent). If
NumTrainingObservations < NumComponents, the
rightmost NumComponents - NumTrainingObservations columns of
Coefficients are 0.
When you specify Coefficients:
You must also specify
Latent.If you also specify
Latent, andVariableWeights, thenCoefficients'*diag(VariableWeights)*Coefficientsmust be orthonormal.If you do not specify
MeansandNumObservations, thenCoefficientsmust be a square matrix.
If you do not specify Coefficients and
Latent, then Coefficients is equal to
zeros(NumPredictors,NumComponents) by default.
Example: Coefficients=0.1*eye(5)
Example: Mdl=incrementalPCA(Coefficients=coeff,Latent=latent,Means=mu,NumObservations=1000)
creates an incremental PCA object Mdl using the outputs returned by
[coeff,~,latent,~,~,mu]=pca(X).
Example: Mdl=incrementalPCA(Coefficients=coeff,Latent=latent)
creates an incremental PCA object Mdl using the outputs returned by
[coeff,latent]=pcacov(V).
Data Types: single | double
This property is read-only.
Principal component variances, namely the eigenvalues of the covariance matrix of
the predictor data, specified as a numeric column vector. The software sets the length
of Latent to NumComponents.
If you specify Latent:
You must also specify
Coefficients.The elements of
Latentmust have non-increasing values.If you do not specify
MeansandNumObservations, the length ofLatentmust equal the number of rows inCoefficients.
The incremental fit function updates
Latent and reorders the columns in descending order by the
principal component variances. If NumTrainingObservations <
NumComponents, the rightmost NumComponents -
NumTrainingObservations columns of Latent are 0.
If you do not specify Latent and
Coefficients when you create IncrementalMdl,
then Latent is equal to zeros(NumComponents,1)
by default.
Example: Latent=0.5*ones(5,1)
Example: Mdl=incrementalPCA(Coefficients=coeff,Latent=latent,Means=mu,NumObservations=1000)
creates an incremental PCA object Mdl using the outputs returned by
[coeff,~,latent,~,~,mu]=pca(X).
Example: Mdl=incrementalPCA(Coefficients=coeff,Latent=latent)
creates an incremental PCA object Mdl using the outputs returned by
[coeff,latent]=pcacov(V).
Data Types: single | double
This property is read-only.
Number of observations processed by the incremental model to estimate
hyperparameters (Mu and Sigma), specified as a
nonnegative integer.
If you specify a positive
EstimationPeriod, and bothStandardizeDataandCenterDataarefalse,incrementalPCAsetsEstimationPeriodto0.If
IncrementalMdlis prepared for incremental PCA (all hyperparameters required for training are specified),incrementalPCAsetsEstimationPeriodto0.If
IncrementalMdlis not prepared for incremental PCA, and eitherStandardizeDataorCenterDataistrue,incrementalPCAsetsEstimationPeriodto1000and estimates the unknown hyperparameters.When processing observations during the estimation period, the software ignores observations that contain at least one missing value.
For more details, see Estimation Period.
Data Types: single | double
This property is read-only.
Percentage of the total variance explained by each principal component, specified as
a numeric column vector. The columns are in descending order by the principal component
variances. The length of ExplainedVariance equals
NumComponents. The values of ExplainedVariance
add up to 100% if NumPredictors and NumComponents
are equal. If NumTrainingObservations <
NumComponents, the last NumComponents -
NumTrainingObservations elements of ExplainedVariance are
0.
You cannot specify ExplainedVariance directly.
Data Types: single | double
This property is read-only.
Flag indicating whether the incremental fit function returns
transformed data, specified as logical 0 (false)
or 1 (true).
If IsWarm is false, the
Xtransformed output of fit consists of
NaN values.
The incremental model IncrementalMdl is warm
(IsWarm is true) if you specify Coefficients
and Latent when
you create IncrementalMdl. Otherwise, IsWarm
becomes true after the incremental fit function
fits the incremental model to WarmupPeriod observations.
If EstimationPeriod >
0, then during the estimation period, fit
does not fit the model and IsWarm is
false.
You cannot specify IsWarm directly.
Data Types: logical
This property is read-only.
Predictor means, specified as a numeric vector.
If Mu is an empty array [] and you specify
CenterData=true or StandardizeData=true, then
the incremental fit function sets Mu to the
predictor variable means estimated during the estimation period specified by EstimationPeriod.
You cannot specify Mu directly. However, if you specify
Means and
NumObservations when you create IncrementalMdl, then
incrementalPCA sets Mu to the values in
Means.
Data Types: single | double
This property is read-only.
Number of principal components to keep after fitting the model, specified as a nonnegative integer.
If you specify NumComponents:
You cannot specify
CoefficientsandLatent.The default value of
NumPredictorsis 0.
If you specify VariableWeights and do not specify NumComponents, then
incrementalPCA sets NumComponents to be equal to
the length of VariableWeights.
If you specify Coefficients
and Latent,
incrementalPCA sets NumComponents to be equal to
the number of rows of Coefficients and the length of
Latent.
Example: NumComponents=3
Data Types: single | double
This property is read-only.
Number of predictor variables used to fit the model, specified as a nonnegative integer.
If you specify NumPredictors:
You cannot specify
CoefficientsandLatent.The default value of
NumComponentsisNumPredictors.NumComponents, if specified, must be less than or equal toNumPredictors.
If you specify VariableWeights, incrementalPCA sets
NumPredictors to be equal to the length of
VariableWeights.
If you specify Coefficients
and Latent,
incrementalPCA sets NumPredictors to be equal to
the number of rows of Coefficients and the length of
Latent.
Data Types: single | double
This property is read-only.
Number of observations fit to the incremental model
IncrementalMdl, specified as a nonnegative numeric scalar.
NumTrainingObservations increases when you pass
IncrementalMdl and training data to fit outside
of the estimation period.
When fitting the model, the software ignores observations that contain at least one missing value.
You cannot specify NumTrainingObservations directly.
Data Types: double
This property is read-only.
Predictor standard deviations, specified as a numeric vector.
If Sigma is an empty array [] and you specify
StandardizeData=true, the incremental fit
function sets Sigma to the predictor variable standard deviations
estimated during the estimation period specified by EstimationPeriod.
You cannot specify Sigma directly.
Data Types: single | double
This property is read-only.
Variable weights, specified as a row vector of positive scalar values.
If you specify VariableWeights, incrementalPCA
sets NumPredictors and NumComponents to be
equal to the length of VariableWeights.
If you specify VariableWeights, Coefficients,
and Latent, then
Coefficients'*diag(VariableWeights)*Coefficients must be
orthonormal.
Example: VariableWeights=0.2*ones(5,1)
Data Types: single | double
This property is read-only.
Number of observations to which the model must be fit before it is
warm, meaning that the incremental fit
function returns transformed data, specified as a nonnegative integer. When processing
observations during the warm-up period, the software ignores observations that contain
at least one missing value.
If
WarmupPeriod<NumComponents,incrementalPCAsetsWarmupPeriodtoNumComponents.If
IncrementalMdlis prepared for incremental PCA (all hyperparameters required for training are specified),incrementalPCAsetsWarmupPeriodto0.If
IncrementalMdlis not prepared for incremental PCA andStandardizeDataistrue,incrementalPCAsetsWarmupPeriodto1000and estimates the unknown hyperparameters.
Data Types: single | double
Object Functions
Examples
Perform principal component analysis (PCA) on an initial data chunk, and then create an incremental PCA model that incorporates the results of the analysis. Fit the incremental model to streaming data and analyze how the model evolves during training.
Load and Preprocess Data
Load the human activity data set.
load humanactivityFor details on the human activity data set, enter Description at the command line.
The data set includes observations containing 60 variables. To simulate streaming data, split the data set into an initial chunk of 1000 observations and a second chunk of 10,000 observations.
Xinitial = feat(1:1000,:); Xstream = feat(1001:11000,:);
Perform Initial PCA
Perform PCA on the initial data chunk by using the pca function. Specify to center the data and keep 10 principal components. Return the principal component coefficients (coeff), principal component variances (latent), and estimated means of the variables (mu).
[coeff,~,latent,~,~,mu]=pca(Xinitial,Centered=true,NumComponents=10);
Create Incremental PCA Model
Create a model for incremental PCA that incorporates the PCA results from the initial data chunk.
IncrementalMdl = incrementalPCA(Coefficients=coeff,Latent=latent, ...
Means=mu,NumObservations=1000);
details(IncrementalMdl) incrementalPCA with properties:
IsWarm: 1
NumTrainingObservations: 0
WarmupPeriod: 0
Mu: [0.7764 0.4931 -0.3407 0.1108 0.0707 0.0485 0.3931 -1.1100 0.0646 0.1703 -1.1020 0.0283 0.0836 -1.0797 0.0139 0.9328 1.2892 1.6731 2.0729 2.5181 2.9511 0.0128 0.0062 0.0039 0.0027 0.0020 0.0016 0.9322 1.3111 … ] (1×60 double)
Sigma: []
ExplainedVariance: [10×1 double]
EstimationPeriod: 0
Latent: [10×1 double]
Coefficients: [60×10 double]
VariableWeights: [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1]
NumComponents: 10
NumPredictors: 60
Methods, Superclasses
IncrementalMdl is an incrementalPCA model object. All its properties are read-only. Because Coefficients and Latent are specified, the model is warm, meaning that the fit function returns transformed observations.
Fit Incremental Model
Fit the incremental model IncrementalMdl to the data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
topEV, the explained variance value of the component with the highest variance, to see how it evolves during incremental fitting.
n = numel(Xstream(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); topEV = zeros(nchunk,1); % Incremental fitting for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); IncrementalMdl = fit(IncrementalMdl,Xstream(ibegin:iend,:)); topEV(j) = IncrementalMdl.ExplainedVariance(1); end
IncrementalMdl is an incrementalPCA model object fitted to all the data in the stream. The fit function fits the model to the data chunk and updates the model properties.
Analyze Incremental Model During Training
Plot the explained variance value of the component with the highest variance to see how it evolves during training.
figure plot(topEV,".-") ylabel("topEV") xlabel("Iteration") xlim([0 nchunk])

The highest explained variance value is 33% after the first iteration, and rapidly rises to 80% after five iterations. The value then gradually approaches 97%.
Create a model for incremental principal component analysis (PCA) and a default incremental linear SVM model for binary classification. Fit the incremental models to streaming data and analyze how the principal components, model parameters, and performance metrics evolve during training. Use the final models to predict activity labels.
Load and Preprocess Data
Load the human activity data set. Randomly shuffle the data.
load humanactivity n = numel(actid); rng(0,"twister") % For reproducibility idx = randsample(n,n); X = feat(idx,:); Y = actid(idx);
For details on the human activity data set, enter Description at the command line.
Responses can be one of five classes: Sitting, Standing, Walking, Running, or Dancing. Dichotomize the response by identifying whether the subject is moving (actid > 2).
Y = Y > 2;
Specify the first 20,000 observations and labels as streaming data, and the remaining observations and labels as test data.
n = 20000; Xstream = X(1:n,:); Ystream = Y(1:n,:); Xtest = X(n+1:end,:); Ytest = Y(n+1:end,:);
Create Incremental Models
Create a model for incremental PCA. Specify to standardize the data, keep 3 principal components, and set a warm-up period of 2000 observations.
IncrementalPCA = incrementalPCA(StandardizeData=true, ...
NumComponents=3,WarmupPeriod=2000);
details(IncrementalPCA) incrementalPCA with properties:
IsWarm: 0
NumTrainingObservations: 0
WarmupPeriod: 2000
Mu: []
Sigma: []
ExplainedVariance: [3×1 double]
EstimationPeriod: 1000
Latent: [3×1 double]
Coefficients: [0×3 double]
VariableWeights: [1×0 double]
NumComponents: 3
NumPredictors: 0
Methods, Superclasses
IncrementalPCA is an incrementalPCA model object. All its properties are read-only. By default, the software sets the hyperparameter estimation period to 1000 observations. The incremental PCA model must be warm (all hyperparameters are estimated) before the fit function returns transformed observations.
Create a default incremental linear SVM model for binary classification by using the incrementalClassificationLinear function.
IncrementalLinear = incrementalClassificationLinear; details(IncrementalLinear)
incrementalClassificationLinear with properties:
Learner: 'svm'
Solver: 'scale-invariant'
BatchSize: 1
Beta: [0×1 double]
Bias: 0
FitBias: 1
FittedLoss: 'hinge'
Lambda: NaN
LearnRate: 1
LearnRateSchedule: 'constant'
Mu: []
Sigma: []
SolverOptions: [1×1 struct]
EstimationPeriod: 0
ClassNames: [0×1 double]
Prior: [1×0 double]
ScoreTransform: 'none'
NumPredictors: 0
NumTrainingObservations: 0
MetricsWarmupPeriod: 1000
MetricsWindowSize: 200
IsWarm: 0
Metrics: [1×2 table]
Methods, Superclasses
IncrementalLinear is an incrementalClassificationLinear model object. All its properties are read-only. IncrementalLinear must be fit to data before you can use it to perform any other operations. By default, the software sets the metrics warm-up period to 1000 observations and the metrics window size to 200 observations.
Fit Incremental Models
Fit the IncrementalPCA and IncrementalLinear models to the streaming data by using the fit and updateMetricsAndFit functions, respectively. To simulate a data stream, fit each model in chunks of 50 observations at a time. At each iteration:
Process 50 observations.
Overwrite the previous incremental PCA model with a new one fitted to the incoming observations.
Return the transformed observations
Xtr.Overwrite the previous incremental classification model with a new one fitted to the incoming transformed observations.
Store , the cumulative metrics, and the window metrics to see how they evolve during incremental learning.
Store
topEV, the explained variance of the component with the highest variance, to see how it evolves during incremental learning.
numObsPerChunk = 50; nchunk = floor(n/numObsPerChunk); ce = array2table(zeros(nchunk,2),"VariableNames",["Cumulative" "Window"]); beta1 = zeros(nchunk,1); topEV = zeros(nchunk,1); % Incremental learning for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); [IncrementalPCA,Xtr] = fit(IncrementalPCA,Xstream(ibegin:iend,:)); IncrementalLinear = updateMetricsAndFit(IncrementalLinear,Xtr, ... Ystream(ibegin:iend)); beta1(j + 1) = IncrementalLinear.Beta(1); ce{j,:} = IncrementalLinear.Metrics{"ClassificationError",:}; topEV(j + 1) = IncrementalPCA.ExplainedVariance(1); end
During the incremental PCA estimation and warm-up periods, the fit function returns the transformed observations as NaNs. After the PCA estimation period and warm-up period, updateMetricsAndFit fits the linear coefficient estimates using the transformed observations. After the metrics warm-up period, IncrementalLinear is warm, and updateMetricsAndFit checks the performance of the model on the incoming transformed observations, and then fits the model to those observations.
Analyze Incremental Models During Training
To see how the highest explained variance, , and performance metrics evolve during training, plot them on separate tiles.
figure t = tiledlayout(3,1); nexttile plot(topEV) ylabel("Top EV [%]") xline(IncrementalPCA.EstimationPeriod/numObsPerChunk,"r-.") xlim([0 nchunk]) ylim([0 100]) nexttile plot(beta1) ylabel("\beta_1") xline((IncrementalPCA.WarmupPeriod+ ... IncrementalPCA.EstimationPeriod)/numObsPerChunk,"b:") xlim([0 nchunk]) nexttile h = plot(ce.Variables); xlim([0 nchunk]) ylabel("Classification Error") xline((IncrementalLinear.MetricsWarmupPeriod+ ... IncrementalPCA.WarmupPeriod+ ... IncrementalPCA.EstimationPeriod)/numObsPerChunk,"g--") legend(h,ce.Properties.VariableNames) xlabel(t,"Iteration")
![Figure contains 3 axes objects. Axes object 1 with ylabel Top EV [%] contains 2 objects of type line, constantline. Axes object 2 with ylabel \beta_1 contains 2 objects of type line, constantline. Axes object 3 with ylabel Classification Error contains 3 objects of type line, constantline. These objects represent Cumulative, Window.](../examples/stats/win64/PerformIncrementalLearningIncorporatingPCAExample_01.png)
The highest explained variance value is 0 during the estimation period and then rapidly rises to 73%. The value then gradually approaches 77%.
The plots suggest that updateMetricsAndFit performs these steps:
Fit after the estimation and warm-up periods only.
Compute the performance metrics after the estimation, warm-up, and metrics warm-up periods only.
Compute the cumulative metrics during each iteration.
Compute the window metrics after processing 200 observations (four iterations).
Predict Activity Labels Using Final Models
Transform the test data using the final incremental PCA model. Predict activity labels for the transformed test data using the final incremental linear classification model.
transformedXtest = transform(IncrementalPCA,Xtest); predictedLabels = predict(IncrementalLinear,transformedXtest);
Create a confusion matrix for the test data.
figure ConfusionTrain = confusionchart(Ytest,predictedLabels);
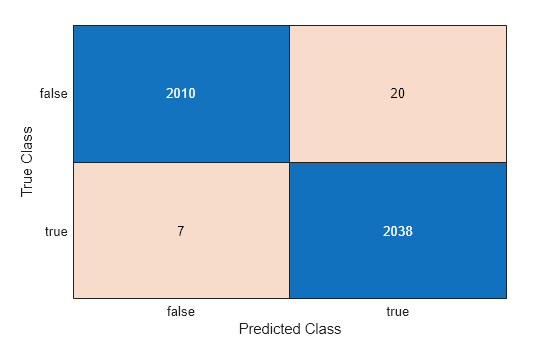
The final model misclassifies only 27 of 4075 observations in the test data.
Tips
You can create an
incrementalPCAmodel object that incorporates the outputs of thepcafunction by using the following code:[coeff,~,latent,~,~,mu]=pca(X); incrementalMdl = incrementalPCA(Coefficients=coeff, ... Latent=latent,Means=mu,NumObservations=1000);You can create an
incrementalPCAmodel object that incorporates the outputs of thepcacovfunction by using the following code:[coeff,latent]=pcacov(V); incrementalMdl = incrementalPCA(Coefficients=coeff,Latent=latent);
Algorithms
During the estimation period, the incremental fit function does not
fit the model. The function uses the first incoming EstimationPeriod observations
to estimate the variable means (Mu) and standard deviations (Sigma). At the end of the
estimation period, the function updates the properties that store the
hyperparameters.
Estimation occurs only when:
EstimationPeriodis positive.IncrementalMdl.MuandIncrementalMdl.Sigmaare empty arrays[].The incremental PCA model object is configured to center or standardize the predictor data (see Center Data and Standardize Data).
Note
If you specify a positive EstimationPeriod and
StandardizeData is false, then
fit resets EstimationPeriod to 0.
When you specify CenterData=true and a positive estimation period
(see Estimation Period), and
IncrementalMdl.Mu and IncrementalMdl.Sigma are
empty, the incremental fit function estimates means using the
estimation period observations.
When you specify StandardizeData=true and a positive estimation
period (see Estimation Period), and
IncrementalMdl.Mu and IncrementalMdl.Sigma are
empty, the incremental fit function estimates means and standard
deviations using the estimation period observations.
When the incremental fit function estimates predictor means and
standard deviations, it computes weighted means and weighted standard deviations using the
estimation period observations. Specifically, the function standardizes predictor
j (xj) using
xj is predictor j, and xjk is observation k of predictor j in the estimation period.
wj is observation weight j.
References
[1] Ross, David A., Jongwoo Lim, Ruei-Sung Lin, and Ming-Hsuan Yang. "Incremental Learning for Robust Visual Tracking." International Journal of Computer Vision 77 (2008): 125-141.
Version History
Introduced in R2024a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)