Skeleton3D
Editor's Note: This file was selected as MATLAB Central Pick of the Week
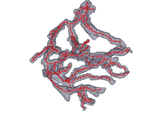
Skeleton3D: Parallel medial axis thinning of a 3D binary volume
This code calculates the 3D medial axis skeleton of an arbitrary 3d binary volume. It is an optimized MATLAB implementation of the homotopic thinning algorithm described in [1]. We developed it to quantify the network of cell processes in bone [2], but it should work on images of any tubular or filamentous structures. An example volume (testvol.mat) is included, along with an example script (Test_Skeleton3D.m). Any comments, corrections or suggestions are highly welcome.
Usage:
skel = Skeleton3D(bin)
where "bin" is a 3D binary image, and "skel" the resulting image containing only the skeleton voxels, or
skel = Skeleton3D(bin,mask)
to mask all foreground voxels in 'mask' from skeletonization, e.g. to preserve certain structures in a image volume.
For additional cleanup, e.g. pruning of short branches, please use my Skel2Graph3D package on MATLAB File Exchange.
This code is inspired by the ITK implementation by Hanno Homann [3] and the Fiji/ImageJ plugin by Ignacio Arganda-Carreras [4]. If you include this in your own work, please cite our original publicaton [2].
Philip Kollmannsberger 09/2013
philipk@gmx.net
References:
[1] Ta-Chih Lee, Rangasami L. Kashyap and Chong-Nam Chu
"Building skeleton models via 3-D medial surface/axis thinning algorithms."
Computer Vision, Graphics, and Image Processing, 56(6):462–478, 1994.
[2] Kollmannsberger, Kerschnitzki et al.,
"The small world of osteocytes: connectomics of the lacuno-canalicular network in bone."
New Journal of Physics 19:073019, 2017.
Cite As
Kollmannsberger, Kerschnitzki et al., "The small world of osteocytes: connectomics of the lacuno-canalicular network in bone." New Journal of Physics 19:073019, 2017.
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Sciences > Biological and Health Sciences > Biomedical Imaging >
- Image Processing and Computer Vision > Computer Vision Toolbox > Recognition, Object Detection, and Semantic Segmentation > Image Category Classification >
Tags
Acknowledgements
Inspired: BiofilmQ, Skel2Graph 3D, Skeleton Length and Aspect Ratio Calculator for Mitochondria
Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
Versions that use the GitHub default branch cannot be downloaded
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.12.0.1 | updated reference |
|
|
| 1.12.0.0 | Fixed title |
|
|
| 1.11.0.0 | Linked to github |
||
| 1.1.0.0 | Removed designation of global variables, and added H1 line for help. For cleaning noise such as short artifacts, please use my Skel2Graph3D package. |
||
| 1.0.0.0 |