cengique/pandora-matlab
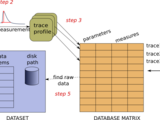
PANDORA is a Matlab Toolbox for analyzing neuronal electrophysiology data and constructing databases.
PANDORA:
Makes database management accessible from your electrophysiology project;
Works offline within Matlab;
Requires no external software;
Is flexible, can easily tie with existing Matlab scripts;
Can query database as in SQL.
Has extensive plotting functionality.
Supports current and voltage clamp protocols for channel fitting.
If you use PANDORA, please cite this paper from your software or publications:
Günay C, Edgerton JR, Li S, Sangrey T, Prinz AA, Jaeger D (2009) Database analysis of simulated and recorded electrophysiological datasets with Pandora's toolbox. Neuroinformatics, 7(2):93-111. doi: 10.1007/s12021-009-9048-z.
You should also cite the RRID: SCR_001831
See the Github for up-to-date information, documentation, and tutorials:
https://github.com/cengique/pandora-matlab
Cite As
Cengiz Gunay (2026). cengique/pandora-matlab (https://github.com/cengique/pandora-matlab), GitHub. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Sciences > Neuroscience > Neuroinformatics >
- Sciences > Neuroscience > Cellular Neuroscience > Electrophysiology >
Tags
Acknowledgements
Inspired by: GODLIKE - A robust single-& multi-objective optimizer, abfload
Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
classes
classes/@chans_db
classes/@cip_trace
classes/@cip_trace_allspikes_profile
classes/@cip_trace_profile
classes/@cip_traces_dataset
classes/@cip_traceset
classes/@cip_traceset_dataset
classes/@cluster_db
classes/@corrcoefs_db
classes/@current_clamp
classes/@dataset_db_bundle
classes/@doc_generate
classes/@doc_multi
classes/@doc_plot
classes/@histogram_db
classes/@histogram_db/private
classes/@mesh_amira
classes/@mesh_amira/private
classes/@model_ct_bundle
classes/@model_data_vcs
classes/@model_ranked_to_physiol_bundle
classes/@params_cip_trace_fileset
classes/@params_results_profile
classes/@params_tests_dataset
classes/@params_tests_db
classes/@params_tests_fileset
classes/@params_tests_profile
classes/@period
classes/@physiol_bundle
classes/@physiol_bundle/private
classes/@physiol_cip_traceset
classes/@physiol_cip_traceset_fileset
classes/@physiol_cip_traceset_fileset/private
classes/@plot_abstract
classes/@plot_bars
classes/@plot_errorbar
classes/@plot_errorbars
classes/@plot_image
classes/@plot_inset
classes/@plot_simple
classes/@plot_stack
classes/@plot_superpose
classes/@ranked_db
classes/@results_profile
classes/@script_array
classes/@script_array_for_cluster
classes/@script_array_loaddb
classes/@script_factory
classes/@spike_shape
classes/@spike_shape_profile
classes/@spikes
classes/@spikes_db
classes/@sql_portal
classes/@stats_db
classes/@tests_3D_db
classes/@tests_db
classes/@tests_db/private
classes/@trace
classes/@trace/private
classes/@trace_allspikes_profile
classes/@trace_profile
classes/@voltage_clamp
classes/@voltage_clamp/private
doc/tutorials/birdsong
examples
functions
test_suite
test_suite/tests_db
Versions that use the GitHub default branch cannot be downloaded
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.4.0.0 | Release 1.4 (https://github.com/cengique/pandora-matlab/releases/tag/v1.4-beta) |
|
|
| 1.3.0.0 | Fixed typo, corrected version, and added link to Wiki. |
|