nfindr
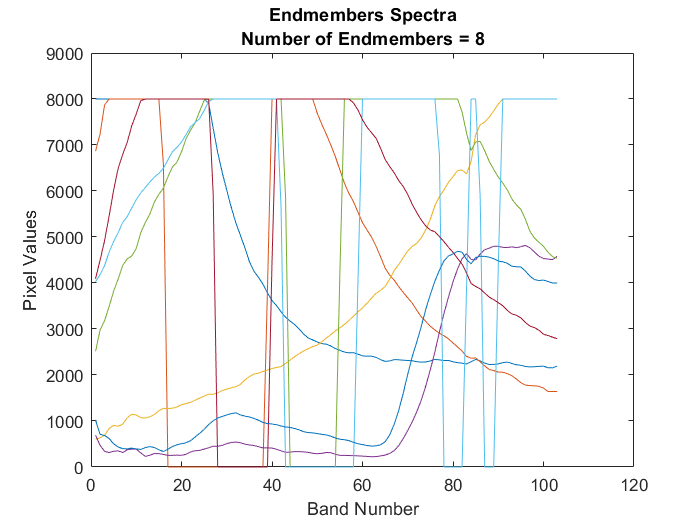
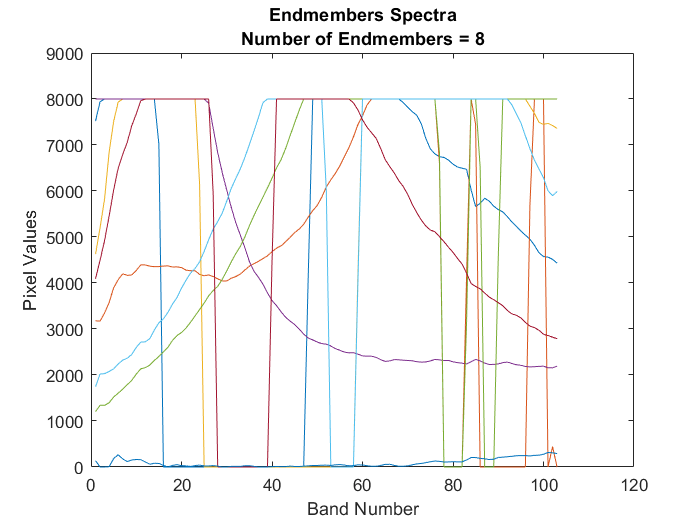
Extract endmember signatures using N-FINDR
Syntax
Description
endmembers = nfindr(inputData,numEndmembers)inputData by
using the N-finder (N-FINDR) algorithm. numEndmembers is the number of
endmember signatures to be extracted using N-FINDR algorithm. For more information about the
N-FINDR method, see Algorithms.
endmembers = nfindr(inputData,numEndmembers,Name=Value)
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
N-FINDR is an iterative approach for finding the endmembers of a hyperspectral data. The method assumes that the volume of a simplex formed by the endmembers (purest pixels) is larger than any other volume defined by any other combination of pixels [1]. The steps involved are as follows:
Compute principal component bands and reduce the spectral dimensionality of the input data by using MNF or PCA. The number of principal component bands to be extracted is set equal to the number of endmembers to be extracted. The endmembers are extracted from the principal component bands.
Randomly select n number of pixel spectra from the reduced data as initial set of endmembers.
For iteration 1, denote the initial set of endmembers as .
Consider the endmembers as vertices of a simplex and compute the volume by using
where .
For iteration 2, Select a new pixel spectra r, such that .
Replace each endmember in the set with r and compute the volume of the simplex V(E(2)).
Replace the ith endmember in the set with r, if the computed volume V(E(2)) is greater than V(E(1)). This results in an updated set of endmembers. For example, if i = 2, the new set of endmembers derived at the end of the second iteration is .
For each iteration, select a new pixel spectra r and repeat steps 5 and 6. Each iteration results in an update set of endmembers. The iteration ends when the total number of iterations reaches the specified value
NumIterations.
References
[1] Winter, Michael E. “N-FINDR: An Algorithm for Fast Autonomous Spectral End-Member Determination in Hyperspectral Data.” Proc. SPIE Imaging Spectrometry V 3753, (October 1999): 266–75. https://doi.org/10.1117/12.366289.
Version History
Introduced in R2020a
See Also
hypercube | ppi | countEndmembersHFC | fippi