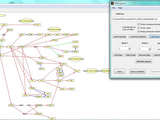
KEGGParser: parsing and editing KEGG pathway maps in Matlab
KEGG pathway database is a collection of manually drawn pathway maps representing current knowledge on molecular interaction and reaction networks, accompanied with KGML (KEGG pathway xml format) files for automatic computational analyses and modeling of metabolic and signaling networks. In a KGML file the pathway is represented as a graph object with entry elements (gene products, compounds, pathways) as its nodes, and relations between elements as edges. However, in most cases there is a lack of correspondence between static images of pathways and accompanying KGML files, so preprocessing of information contained in a KGML file is needed before it can be used in automatic analysis. Several KGML parsers have been developed recently, both standalone or integrated in different bioinformatics packages.
We introduce KEGGParser (pathway parser/editor), which is based on Matlab biograph class.
Features:
1. Retrieval, parsing and automatic correction for protein-compound-protein interactions, group nodes and binding directions;
2. Pathway graph editing (edge and node manipulations) based on "Graph manipulation" (http://www.mathworks.com/matlabcentral/fileexchange/37475-graph-manipulation);
3. Analysis of parsed pathways can be performed using Matlab built-in graph-based calculations.
See more details in Arakelyan A, Nersisyan L.KEGGParser: parsing and editing KEGG pathway maps in Matlab. Bioinformatics. 2013 Feb 15;29(4):518-9. doi: 10.1093/bioinformatics/bts730.
Cite As
Arsen Arakelyan (2025). KEGGParser: parsing and editing KEGG pathway maps in Matlab (https://www.mathworks.com/matlabcentral/fileexchange/37561-keggparser-parsing-and-editing-kegg-pathway-maps-in-matlab), MATLAB Central File Exchange. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Industries > Biotech and Pharmaceutical > Genomics and Next Generation Sequencing >
- Computational Biology > Bioinformatics Toolbox >
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.9.0.0 | 1. Minor bugs are fixed.
|
||
| 1.8.0.0 | Recent updates
2. parse_KEGG_xml.m updated to allow creation of graphs without edges.
|
||
| 1.7.0.0 | Fixed problem with parsing relations without subtypes. Added automatic correction for protein-compound-protein interactions, group nodes and binding directions. |
||
| 1.6.0.0 | Fixes:
|
||
| 1.5.0.0 | I have found a bug, that results in wrong assignment of interaction types. It is fixed. Scilab version of KEGGParser is also included. Works with Scilab 5.4+metanet |
||
| 1.0.0.0 |