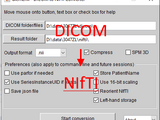
xiangruili/dicm2nii
Editor's Note: This file was selected as MATLAB Central Pick of the Week
Cite As
Xiangrui Li (2025). xiangruili/dicm2nii (https://github.com/xiangruili/dicm2nii/releases/tag/v2023.02.23), GitHub. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxCategories
- Image Processing and Computer Vision > Image Processing Toolbox > Import, Export, and Conversion >
- Sciences > Neuroscience > Human Brain Mapping > Neuroimaging >
- Image Processing and Computer Vision > Image Processing Toolbox > Get Started with Image Processing Toolbox >
- Industries > Medical Devices > DICOM Format >
Tags
Acknowledgements
Inspired: MRIqual, DICOM to any Image Format Converter, Medical Image Reader and Viewer
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
Versions that use the GitHub default branch cannot be downloaded
| Version | Published | Release Notes | |
|---|---|---|---|
| 2023.2.23.0 | See release notes for this release on GitHub: https://github.com/xiangruili/dicm2nii/releases/tag/v2023.02.23 |
||
| 2022.03.06 | update version |
|
|
| 2018.08.08 | 1. use GetFullPath from Jan
|
|
|
| 2018.6.14.0 | dicm2nii: implement 16-bit range scale (default off);
|
|
|
| 2018.6.5.0 | 1. dicm_hdr: can read Philips xml file; many improvements for PAR dealing
|
||
| 2018.5.23.0 | dicm2nii: take care of some special Philips cases:
|
||
| 2018.4.20.0 | 1. dicm2nii: bug fix for long file name
|
||
| 2018.3.9.0 | nii_viewer:
|
||
| 2018.1.19.0 | 1. nii_viewer: map inflated surface click to correct location in volume, even for old matlab
|
||
| 2018.1.3.0 | nii_viewer: support inflated surface to volume mapping. Tested only for HCP surfaces. |
||
| 2017.12.29.0 | surface view uses combined overlay, so it works smoothly for different environment. |
||
| 2017.12.28.0 | 1. nii_viewer: implement surface view for CIfTI (HCP gii template included).
|
||
| 2017.12.14.0 | 1. dicm2ii: work for Siemens multiframe dicom.
|
||
| 2017.10.31.0 | 1. nii_view: two more layout including compact one-row display
|
||
| 2017.9.27.0 | 1. Save VolumeTiming for missing volumes based on BIDS;
|
||
| 2017.8.3.0 | 1. Bug fix for reading Philips PAR file;
|
||
| 2017.7.20.0 | 1. Add functionSignatures.json file for tab auto-completion;
|
||
| 2017.6.18.0 | 1. dicm2nii & nii_viewer: can read big-endian AFNI image;
|
||
| 2017.3.22.0 | 1. dicm2nii: better precision requirement for ImagePositionPatient;
|
||
| 2017.2.15.0 | 1. nii_viewer: Can open DICOM etc files by converting into NIfTI without saving, allowing to view image from one or more files without converting.
|
||
| 2017.1.3.0 | anonymize_dicm: replace PatientName with ID rather than scratching it. |
||
| 2016.12.29.0 | 1. nii_moco: new implementation to perform motion correction;
|
||
| 2016.11.30.0 | 1. dicm2nii: fix for irregular image order in Philips multi-frame dicom;
|
||
| 2016.11.15.0 | 1. nii_viewer: allow to manually remove excessive neck tissue;
|
||
| 2016.10.10.0 | 1. nii_viewer: implement warped overlay;
|
||
| 2016.9.21.0 | dicm2nii: fix bug introduced by update hours ago (slope/intercept wrongly applied to files expect 1st). |
||
| 2016.9.20.0 | 1. dicm2nii: Add preferences to GUI
|
||
| 2016.8.26.0 | 1. dicm2nii: add pref to allow to not use SeriesInstanceUID;
|
||
| 2016.7.10.0 | nii_viewer: file list GUI improvement; implement histogram plot and sphere ROI creation.
|
||
| 2016.6.10.0 | nii_viewer: add 'RGB' LUT so 3-volume image can be in RGB;
|
||
| 2016.6.1.0 | dicm2nii: always save ReadoutSeconds for topup;
|
||
| 2016.5.21.0 | nii_stc: new file for nifti slice timing correction.
|
||
| 2016.5.16.0 | dicm2nii: big fix for error due to missing file(s). |
||
| 2016.5.12.0 | dicm2nii:
|
||
| 2016.5.9.0 | nii_viewer: Update phase LUTs to three; Add "Open in new window" |
||
| 2016.5.6.0 | Corrected version number |
||
| 2016.5.5.0 | dicm2nii: better mosaic detection for old/bad data; Convert MoCo series by default.
|
||
| 2016.3.24.0 | 1. dicm2nii: Fix Siemens mosaic detection for rare cases.
|
||
| 2016.1.31.0 | 1. nii_viewer: bug fix for messed-up callback in set_file;
|
||
| 2016.1.27.0 | 1. dicm2nii: support big endian dicom files;
|
||
| 2016.1.15.0 | check update: fix the problem to download into current directory. |
||
| 2016.1.14.0 | 1. nii_viewer: implement time course plot;
|
||
| 2016.1.12.0 | 1. Implement update check from dicm2nii and nii_viewer GUI;
|
||
| 2016.1.5.0 | Fixes and improvements for dicm2nii, nii_tool, and mostly for nii_viewer:
|
||
| 2015.11.28.0 | nii_viewer: bug fix introduced in previous update (reorient problem) |
||
| 2015.11.27.0 | nii_viewer:
|
||
| 2015.11.22.0 | nii_viewer fix and improvement:
|
||
| 2015.11.20.0 | 1. dicm2nii: GUI supports drag and drop of folder/files; optionally save JSON file;
|
||
| 2015.11.5.0 | Bug fix for Show NIfTI hdr/ext |
||
| 2015.11.4.0 | 1. Implement nii_viewer.m for nii visualization;
|
||
| 2015.9.26.0 | 1. Take care irregular slice ordering for PAR/REC and multiframe dicom;
|
||
| 1.44.0.0 | 1. nii_tool: auto detects RGB style for image reading;
|
||
| 1.43.0.0 | Major: Store text NIfTI extension for information, like DTI bval/bvec, slice timing, unwarp parameters etc
|
||
| 1.42.0.0 | Do not store ecode=40 NIfTI extension due to possible problem with FSL |
||
| 1.41.0.0 | Fix problems with latest Matlab versions: uint32 figure handle and parpool. |
||
| 1.40.0.0 | 1. Can convert BrainVoyager files into NIfTI;
|
||
| 1.39.0.0 | 1. Works multi subject/study data;
|
||
| 1.38.0.0 | 1. Bug fix for phase encoding direction;
|
||
| 1.37.0.0 | 1. dicm2nii can save SPM style NIfTI;
|
||
| 1.36.0.0 | 1. Include new file nii_tool.m, which can read/write almost any datatype of NIfTI files with different versions;
|
||
| 1.34.0.0 | 1. Fix error for multiframe dicom with single frame;
|
||
| 1.33.0.0 | 1. Use ImagePositionPatient to derive SliceThickness if possible;
|
||
| 1.32.0.0 | See previous update |
||
| 1.31.0.0 | 1. Use parallel tool if available and worthy;
|
||
| 1.30.0.0 | 1. bug fix for missing number of slices in GE multiframe dicom;
|
||
| 1.29.0.0 | Optional output argument for sort_dicm |
||
| 1.28.0.0 | 1. Include new file sort_dicm, which sorts files for different subjects into sub-folders;
|
||
| 1.27.0.0 | Take care of different carriage return issue in Philips PAR file |
||
| 1.26.0.0 | 1. Take care of missing VR by some 3rd party dicom converter;
|
||
| 1.25.0.0 | 1. Fix the problem caused by PAR 4.2 keyword case change;
|
||
| 1.24.0.0 | 1. can deal with compressed tgz, tar file
|
||
| 1.23.0.0 | 1. Take care of non-unique ixyz;
|
||
| 1.22.0.0 | 1. Image storage change to make DTI data work for FSL 5.05 and later
|
||
| 1.21.0.0 | 1. Fix to work for new GE data, including DTI;
|
||
| 1.20.0.0 | Not set cal_min and cal_max anymore, so avoid display range problem. |
||
| 1.19.0.0 | 1. Store FSL slice timing in dcmHeaders.mat;
|
||
| 1.18.0.0 | 1. bug fix for MoCo series detection;
|
||
| 1.17.0.0 | 1. Support dicom without meta info;
|
||
| 1.16.0.0 | Majors:
|
||
| 1.15.0.0 | 1. Bug fix and improvement for DTI parameters.
|
||
| 1.14.0.0 | 1. Implement conversion for AFNI HEAD/BRIK files.
|
||
| 1.13.0.0 | 1. Minor improvement for dicm_hdr: read V4.1 PAR file correctly;
|
||
| 1.12.0.0 | 1. Support implicit VR dicom;
|
||
| 1.11.0.0 | 1. First working version for Philips multiframe dicom
|
||
| 1.10.0.0 | 1. Works for Siemens non-mosaic DTI data too.
|
||
| 1.9.0.0 | Major: first implementation for conversion of GE and Philips data.
|
||
| 1.8.0.0 | Bug fix for DTI bval/bvec files introduced in last update |
||
| 1.6.0.0 | 1. Try and suggest to use pigz for gz compression
|
||
| 1.5.0.0 | A bug fix for '*' input, and some minor improvement for dicom header reading |
||
| 1.4.0.0 | Some SPM users may not have Imaging Processing Toolbox. This update removed this dependency. This also speeds up the dicom header parsing. |
||
| 1.3.0.0 | 1. File name is changed from dcm2nii.m to dicm2nii.m. This will avoid confusion with MRICron's dcm2nii converter.
|
||
| 1.2.0.0 | 1. Allow to specify a subjectID for nii;
|
||
| 1.1.0.0 | Some minor improvement for the code. Add a line for usage. |
||
| 1.0.0.0 |