Brainstorm: A MATLAB Based, Open-Source Application for Advanced MEG/EEG Data Processing and Visualization
By François Tadel and Sylvain Baillet, McConnell Brain Imaging Centre, McGill University; John C. Mosher, Cleveland Clinic Epilepsy Center, Case Western Reserve University; and Richard M. Leahy, University of Southern California
The sight of a snake-like object near your feet sends out neural signals to the amygdala, a rather archaic brain structure, which triggers a rapid fight-or-flight response. It is not until tens of milliseconds later that the neocortex, which evolved more recently, performs a more detailed analysis of the same sensory input and determines that the object is a harmless garden hose.
Brain imaging, or neuroimaging, provides neuropsychologists and neuroscientists with noninvasive techniques for studying brain processes like fight-or-flight. This multidisciplinary field of science has developed rapidly since the late 1990s. Functional magnetic resonance imaging (fMRI) can be used to map the topography of brain activity, but with a temporal resolution of about one second, it is not as useful for studying the dynamic flow of operations performed by the brain. Modern electroencephalography (EEG) and magnetoencephalography (MEG) technology have millisecond temporal resolution and enable researchers to track neural activity at the natural speed of the brain. The other benefit of MEG and EEG is their sensitivity to neural electrochemical signaling, enabling hypotheses testing of the electrophysiological mechanisms of regional brain activity. Finally, the high temporal resolution and synoptic coverage of cortical activity provided by MEG and EEG reveal patterns of functional connectivity between brain regions.
The emergence of functional connectivity in neuroimaging is extending research beyond the mapping of essential functions such as perception, motor control, memory, and language. The current view is that most brain functions require the coordination of activity in multiple brain regions that assemble dynamically into networks. The millisecond temporal resolution of MEG and EEG is currently the best asset for neuroscientists to capture the complexity of electrophysiological activity emerging from many brain regions simultaneously. Many neuroscientists also consider that most neurological and psychiatric conditions may be associated with altered forms of functional connectivity, such as those causing seizure in the epileptic brain. This transformative hypothesis is expected to substantially increase our understanding of the brain and enable clinicians to prevent and treat a vast and complex spectrum of neurological disorders.
MEG/EEG instruments capture electrical activity from the entire volume of the brain at a typical rate of 1,000 times per second, yielding roughly 100 MB of data per minute. Visualizing and processing the vast amounts of data these instruments generate is a significant challenge for neuroscientists. Compounding this challenge is a relative disconnect between the neuroscience and neuropsychology communities and the imaging scientists who develop methods for time-resolved image reconstruction from MEG and EEG data. These physicists and electrical engineers typically develop advanced and highly technical tools, which they find hard or time-consuming to translate to the neuroscience researchers.
To resolve this disconnect, we created Brainstorm, an open-source software application developed with MATLAB®. Brainstorm enables neuroscience researchers with no programming experience to visualize and process large volumes of MEG and EEG data (Figure 1).
Over the past ten years, Brainstorm has been downloaded by over ten thousand researchers worldwide and cited in more than 250 journal articles, including such influential journals as Nature Neuroscience, Science, and the Proceedings of the National Academy of Sciences. The Brainstorm user community consists of approximately 70% neuroscientists (clinical scientists, neuropsychologists, and cognitive neuroscientists), with the imaging scientists and engineers who specialize in MEG/EEG method development making up the remaining 30%. Brainstorm is also frequently used in other research environments—for example, in studying the pathological brain activity of patients with epilepsy, and in small animal electrophysiology.
Why MATLAB?
MATLAB offers several advantages over traditional programming languages. First, we are physicists and engineers, and when we began to develop Brainstorm, we knew little about software development. MATLAB made it easy for us to develop a sophisticated scientific application with an extensive graphical interface. Second, MATLAB is widely used in the scientific community. As a result, researchers can interact directly with their data using Brainstorm, contribute new plug-ins, and exchange ideas and code prototypes with other Brainstorm users.
Third, MATLAB is a productive development environment. The ability to debug interactively and visualize data in MATLAB enables us to rapidly prototype new capabilities for Brainstorm. Brainstorm’s interface is written in Java embedded in MATLAB scripts, taking advantage of the tight integration between the two languages. For most of the 2D and 3D data figures, Brainstorm uses the MATLAB graphic system, which simplifies the building of complex and interactive scenes.
Brainstorm users can download and install the software even if they are not licensed MATLAB users; with MATLAB Compiler™ and MATLAB Builder™ JA, we provide a standalone version of the software that runs on Windows, Linux, and Mac OS X systems.
For future developments, we consider a key feature is the closer integration with the Python environment that was added in MATLAB 2014b. The popularity of the open-source Python language is growing rapidly in academia, especially among new trainees in scientific computing. This new generation of students and researchers produces a great diversity of Python scripts and libraries, which MATLAB users can benefit from. Our opinion is that it is essential and mutually beneficial to continue to nurture and develop the connection between MATLAB and the open-source and open-environment community.
The Brainstorm Research Workflow
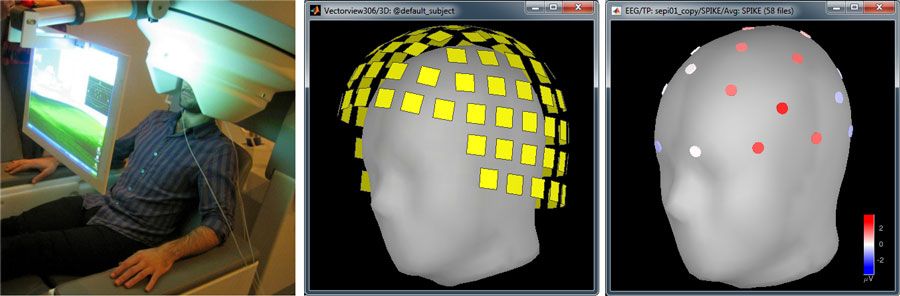
In a typical MEG and EEG study, between 1 and 100 (typically, 15) subjects are tested in multiple experimental conditions. EEG data is collected from electrodes attached to the subject’s scalp, and MEG data is collected via sensors located inside the MEG rigid helmet (Figure 2).
To analyze the results of these experiments, researchers use multimodal datasets: the MEG or simultaneous EEG recordings, the 3D locations of the sensors, and the anatomy of the subject’s head volume—the latter typically obtained with MRI.
In a standard Brainstorm workflow, the first step is to import the MRI data along with the surfaces representing the cerebral cortex and scalp. The MEG/EEG space is coregistered with the MRI by aligning the sensors on the scalp surface of the subject.
Next, the MEG/EEG recordings are reviewed and preprocessed. Brainstorm’s data viewer allows users to browse efficiently through long recordings, adjust the gain of traces, mark events of interest, and tag bad segments and noisy sensors and other interferences. (Figure 3). Because there is no standard file format for MEG/EEG data, we have developed vendor-specific data-parsing routines in MATLAB for the most common formats. We continue to add support for new formats as they emerge from our diverse user community.
Preprocessing raw data mainly involves removing data components caused by noise sources and artifacts. MEG/EEG signals are typically contaminated by noise from the acquisition system itself (sensor noise), the environment (such as 50Hz/60Hz power lines, stimulation devices, and building vibrations) and from the subject (movements, eye blinks, heartbeats, and breathing). Depending on the type and amount of noise, a combination of frequency filters and spatial projectors is an effective noise-reduction approach.
After preprocessing, researchers import data segments of interest into the Brainstorm database, which creates a set of .MAT files that store structured MATLAB variables of multiple types. In a typical event-related setup, the data is organized into epoch segments surrounding stimulus events, such as sounds or images presented to the subject.
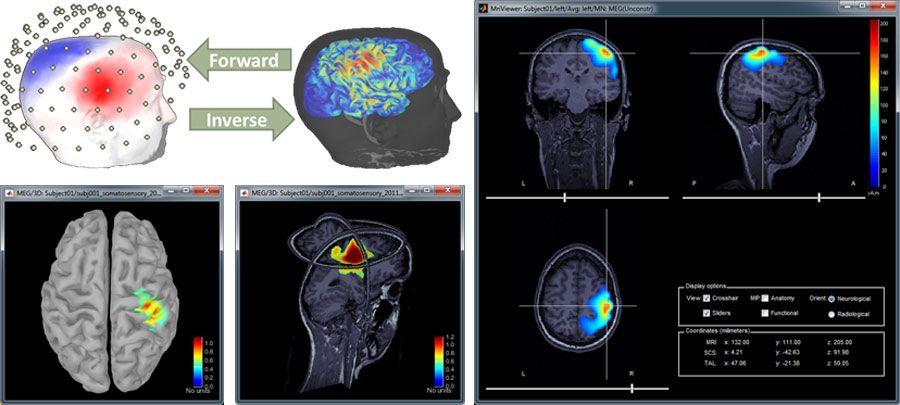
The researcher then begins reconstructing brain activity from the measured data. In this phase, Brainstorm uses forward modeling with boundary element methods (BEMs) to compute a head model by mapping neural currents and MEG/EEG sensor measurements. The researcher then uses Brainstorm’s advanced inverse modeling capabilities to locate the cortical sources that produced a specific set of MEG/EEG recordings (Figure 4).
Once neuronal signals are accessible at both the sensor and source levels, more advanced analysis can be performed to explore effects specific to the experimental task. Spectral and time-frequency decompositions, measures of functional connectivity between brain regions, cross-frequency coupling mechanisms, and statistical inference are typical families of methods used with time-resolved brain images and signals inside Brainstorm (Figure 5).

All Brainstorm features are accessible using standard keyboard and mouse inputs. After prototyping the analysis pipeline on a few datasets using the graphical interface, researchers typically need to repeat the same operations for tens or hundreds of similar datasets. Brainstorm offers a flexible graphic batching tool for this purpose: the user selects the files to process from a database explorer, and lists all the operations to be applied sequentially on selected files (Figure 6), and Brainstorm automatically generates a MATLAB batch script. This script can be executed as is, kept as a research log, or customized with the addition of operations not available in Brainstorm.
Engaging the Brainstorm Development Community
Because Brainstorm is open and extensible, users are invited to contribute their own MATLAB code to replace or complement most existing Brainstorm operations, including filtering, statistical analysis, signal and image processing, forward modeling, and inverse modeling. In addition to sharing these plug-ins, researchers support the open-source development effort by contributing their own ideas and MATLAB code. In some cases we optimize the contributed code before incorporating it into Brainstorm, but a number of experienced MATLAB programmers have contributed code that we merged into Brainstorm virtually untouched.
The training courses that we conduct worldwide are an additional source of new ideas. In the past year, we have organized training sessions for about 900 users at sites in Europe, Asia, North America, and the Middle East. These sessions are an excellent way for new users to learn Brainstorm, complementing our comprehensive online tutorials and training datasets. Just as importantly, the training sessions offer an unparalleled opportunity for us to meet our users. We learn about their needs, their research, and any aspects of Brainstorm they find confusing or difficult. Based on this input, we use MATLAB to prototype improvements for incorporation into a future Brainstorm release.
We are always improving Brainstorm. Future development in MATLAB will enhance support for combining multiple imaging modalities and add more processing methods to keep up with cutting-edge published research. Researchers will then be able to use Brainstorm to visualize and analyze not only MEG/EEG data but also optical imaging (fNIRS), functional MRI, diffusion weighted MRI, CT scans, and positron emission tomography (PET) data in a single environment.
Acknowledgement
Research and software reported in this publication was supported by the Montreal Neurological Institute, and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number R01EB009048. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Published 2015 - 92265v00