estimate
Perform predictor variable selection for Bayesian linear regression models
Syntax
Description
To estimate the posterior distribution of a standard Bayesian linear regression
model, see estimate.
PosteriorMdl = estimate(PriorMdl,X,y)estimate also performs predictor variable selection.
PriorMdl specifies the joint prior distribution of the parameters,
the structure of the linear regression model, and the variable selection algorithm.
X is the predictor data and y is the response
data. PriorMdl and PosteriorMdl are not the same
object type.
To produce PosteriorMdl, estimate updates the
prior distribution with information about the parameters that it obtains from the
data.
NaNs in the data indicate missing values, which
estimate removes using list-wise deletion.
PosteriorMdl = estimate(PriorMdl,X,y,Name,Value)'Lambda',0.5 specifies that the shrinkage parameter value for
Bayesian lasso regression is 0.5 for all coefficients except the
intercept.
If you specify Beta or Sigma2, then
PosteriorMdl and PriorMdl are equal.
[
uses any of the input argument combinations in the previous syntaxes and also returns a
table that includes the following for each parameter: posterior estimates, standard
errors, 95% credible intervals, and posterior probability that the parameter is greater
than 0.PosteriorMdl,Summary]
= estimate(___)
Examples
Consider the multiple linear regression model that predicts US real gross national product (GNPR) using a linear combination of industrial production index (IPI), total employment (E), and real wages (WR).
For all , is a series of independent Gaussian disturbances with a mean of 0 and variance .
Assume the prior distributions are:
For k = 0,...,3, has a Laplace distribution with a mean of 0 and a scale of , where is the shrinkage parameter. The coefficients are conditionally independent.
. and are the shape and scale, respectively, of an inverse gamma distribution.
Create a prior model for Bayesian lasso regression. Specify the number of predictors, the prior model type, and variable names. Specify these shrinkages:
0.01for the intercept10forIPIandWR1e5forEbecause it has a scale that is several orders of magnitude larger than the other variables
The order of the shrinkages follows the order of the specified variable names, but the first element is the shrinkage of the intercept.
p = 3; PriorMdl = bayeslm(p,'ModelType','lasso','Lambda',[0.01; 10; 1e5; 10],... 'VarNames',["IPI" "E" "WR"]);
PriorMdl is a lassoblm Bayesian linear regression model object representing the prior distribution of the regression coefficients and disturbance variance.
Load the Nelson-Plosser data set. Create variables for the response and predictor series.
load Data_NelsonPlosser X = DataTable{:,PriorMdl.VarNames(2:end)}; y = DataTable{:,"GNPR"};
Perform Bayesian lasso regression by passing the prior model and data to estimate, that is, by estimating the posterior distribution of and . Bayesian lasso regression uses Markov chain Monte Carlo (MCMC) to sample from the posterior. For reproducibility, set a random seed.
rng(1); PosteriorMdl = estimate(PriorMdl,X,y);
Method: lasso MCMC sampling with 10000 draws
Number of observations: 62
Number of predictors: 4
| Mean Std CI95 Positive Distribution
-------------------------------------------------------------------------
Intercept | -1.3472 6.8160 [-15.169, 11.590] 0.427 Empirical
IPI | 4.4755 0.1646 [ 4.157, 4.799] 1.000 Empirical
E | 0.0001 0.0002 [-0.000, 0.000] 0.796 Empirical
WR | 3.1610 0.3136 [ 2.538, 3.760] 1.000 Empirical
Sigma2 | 60.1452 11.1180 [42.319, 85.085] 1.000 Empirical
PosteriorMdl is an empiricalblm model object that stores draws from the posterior distributions of and given the data. estimate displays a summary of the marginal posterior distributions in the MATLAB® command line. Rows of the summary correspond to regression coefficients and the disturbance variance, and columns correspond to characteristics of the posterior distribution. The characteristics include:
CI95, which contains the 95% Bayesian equitailed credible intervals for the parameters. For example, the posterior probability that the regression coefficient ofIPIis in [4.157, 4.799] is 0.95.Positive, which contains the posterior probability that the parameter is greater than 0. For example, the probability that the intercept is greater than 0 is0.427.
Plot the posterior distributions.
plot(PosteriorMdl)
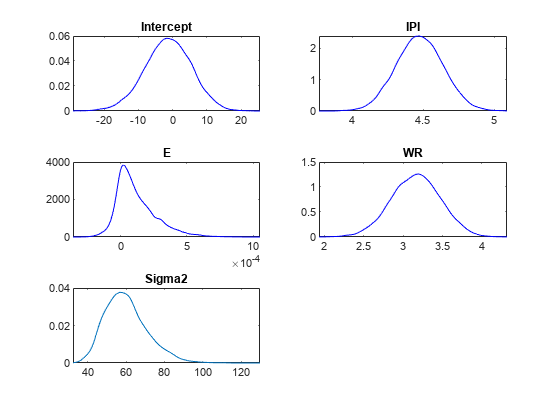
Given the shrinkages, the distribution of E is fairly dense around 0. Therefore, E might not be an important predictor.
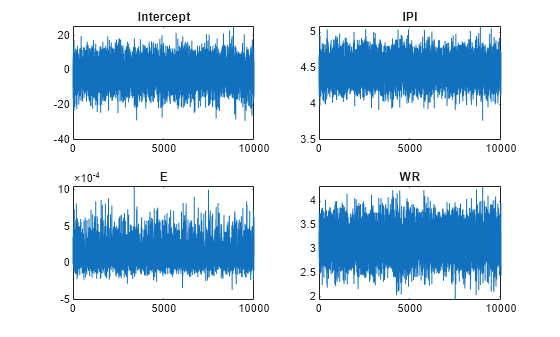
By default, estimate draws and discards a burn-in sample of size 5000. However, a good practice is to inspect a trace plot of the draws for adequate mixing and lack of transience. Plot a trace plot of the draws for each parameter. You can access the draws that compose the distribution (the properties BetaDraws and Sigma2Draws) using dot notation.
figure; for j = 1:(p + 1) subplot(2,2,j); plot(PosteriorMdl.BetaDraws(j,:)); title(sprintf('%s',PosteriorMdl.VarNames{j})); end
figure;
plot(PosteriorMdl.Sigma2Draws);
title('Sigma2');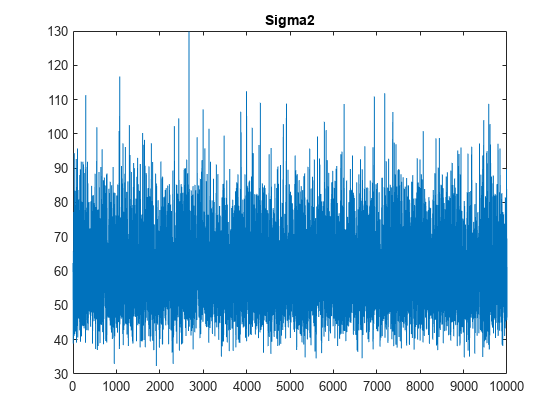
The trace plots indicate that the draws seem to mix well. The plots show no detectable transience or serial correlation, and the draws do not jump between states.
Consider the regression model in Select Variables Using Bayesian Lasso Regression.
Create a prior model for performing stochastic search variable selection (SSVS). Assume that and are dependent (a conjugate mixture model). Specify the number of predictors p and the names of the regression coefficients.
p = 3; PriorMdl = mixconjugateblm(p,'VarNames',["IPI" "E" "WR"]);
Load the Nelson-Plosser data set. Create variables for the response and predictor series.
load Data_NelsonPlosser X = DataTable{:,PriorMdl.VarNames(2:end)}; y = DataTable{:,'GNPR'};
Implement SSVS by estimating the marginal posterior distributions of and . Because SSVS uses Markov chain Monte Carlo for estimation, set a random number seed to reproduce the results.
rng(1); PosteriorMdl = estimate(PriorMdl,X,y);
Method: MCMC sampling with 10000 draws
Number of observations: 62
Number of predictors: 4
| Mean Std CI95 Positive Distribution Regime
----------------------------------------------------------------------------------
Intercept | -18.8333 10.1851 [-36.965, 0.716] 0.037 Empirical 0.8806
IPI | 4.4554 0.1543 [ 4.165, 4.764] 1.000 Empirical 0.4545
E | 0.0010 0.0004 [ 0.000, 0.002] 0.997 Empirical 0.0925
WR | 2.4686 0.3615 [ 1.766, 3.197] 1.000 Empirical 0.1734
Sigma2 | 47.7557 8.6551 [33.858, 66.875] 1.000 Empirical NaN
PosteriorMdl is an empiricalblm model object that stores draws from the posterior distributions of and given the data. estimate displays a summary of the marginal posterior distributions in the command line. Rows of the summary correspond to regression coefficients and the disturbance variance, and columns correspond to characteristics of the posterior distribution. The characteristics include:
CI95, which contains the 95% Bayesian equitailed credible intervals for the parameters. For example, the posterior probability that the regression coefficient ofE(standardized) is in [0.000, 0.0.002] is 0.95.Regime, which contains the marginal posterior probability of variable inclusion ( for a variable). For example, the posterior probabilityEthat should be included in the model is 0.0925.
Assuming that variables with Regime < 0.1 should be removed from the model, the results suggest that you can exclude the unemployment rate from the model.
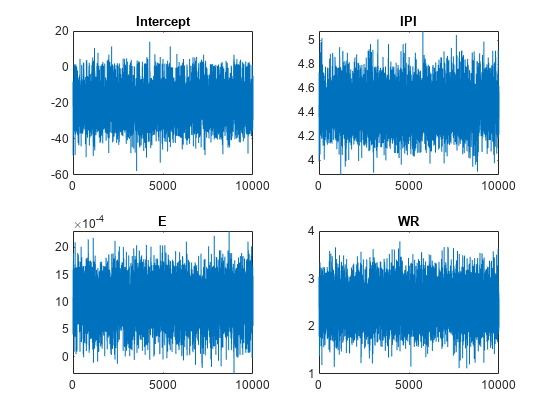
By default, estimate draws and discards a burn-in sample of size 5000. However, a good practice is to inspect a trace plot of the draws for adequate mixing and lack of transience. Plot a trace plot of the draws for each parameter. You can access the draws that compose the distribution (the properties BetaDraws and Sigma2Draws) using dot notation.
figure; for j = 1:(p + 1) subplot(2,2,j); plot(PosteriorMdl.BetaDraws(j,:)); title(sprintf('%s',PosteriorMdl.VarNames{j})); end
figure;
plot(PosteriorMdl.Sigma2Draws);
title('Sigma2');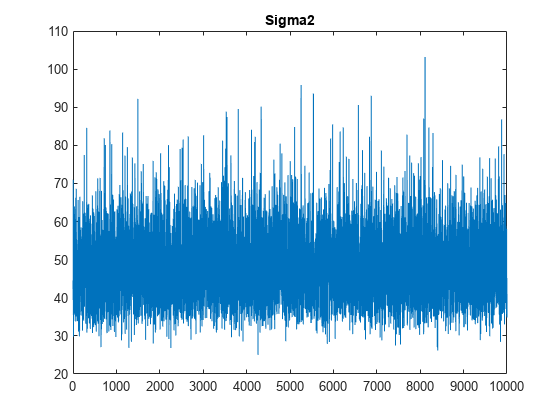
The trace plots indicate that the draws seem to mix well. The plots show no detectable transience or serial correlation, and the draws do not jump between states.
Consider the regression model and prior distribution in Select Variables Using Bayesian Lasso Regression.
Create a Bayesian lasso regression prior model for 3 predictors and specify variable names. Specify the shrinkage values 0.01, 10, 1e5, and 10 for the intercept, and the coefficients of IPI, E, and WR.
p = 3; PriorMdl = bayeslm(p,'ModelType','lasso','VarNames',["IPI" "E" "WR"],... 'Lambda',[0.01; 10; 1e5; 10]);
Load the Nelson-Plosser data set. Create variables for the response and predictor series.
load Data_NelsonPlosser X = DataTable{:,PriorMdl.VarNames(2:end)}; y = DataTable{:,"GNPR"};
Estimate the conditional posterior distribution of given the data and that , and return the estimation summary table to access the estimates.
rng(1); % For reproducibility [Mdl,SummaryBeta] = estimate(PriorMdl,X,y,'Sigma2',10);
Method: lasso MCMC sampling with 10000 draws
Conditional variable: Sigma2 fixed at 10
Number of observations: 62
Number of predictors: 4
| Mean Std CI95 Positive Distribution
------------------------------------------------------------------------
Intercept | -8.0643 4.1992 [-16.384, 0.018] 0.025 Empirical
IPI | 4.4454 0.0679 [ 4.312, 4.578] 1.000 Empirical
E | 0.0004 0.0002 [ 0.000, 0.001] 0.999 Empirical
WR | 2.9792 0.1672 [ 2.651, 3.305] 1.000 Empirical
Sigma2 | 10 0 [10.000, 10.000] 1.000 Empirical
estimate displays a summary of the conditional posterior distribution of . Because is fixed at 10 during estimation, inferences on it are trivial.
Display Mdl.
Mdl
Mdl =
lassoblm with properties:
NumPredictors: 3
Intercept: 1
VarNames: {4×1 cell}
Lambda: [4×1 double]
A: 3
B: 1
| Mean Std CI95 Positive Distribution
---------------------------------------------------------------------------
Intercept | 0 100 [-200.000, 200.000] 0.500 Scale mixture
IPI | 0 0.1000 [-0.200, 0.200] 0.500 Scale mixture
E | 0 0.0000 [-0.000, 0.000] 0.500 Scale mixture
WR | 0 0.1000 [-0.200, 0.200] 0.500 Scale mixture
Sigma2 | 0.5000 0.5000 [ 0.138, 1.616] 1.000 IG(3.00, 1)
Because estimate computes the conditional posterior distribution, it returns the model input PriorMdl, not the conditional posterior, in the first position of the output argument list.
Display the estimation summary table.
SummaryBeta
SummaryBeta=5×6 table
Mean Std CI95 Positive Distribution Covariances
__________ __________ ________________________ ________ _____________ _______________________________________________________________________
Intercept -8.0643 4.1992 -16.384 0.01837 0.0254 {'Empirical'} 17.633 0.17621 -0.00053724 0.11705 0
IPI 4.4454 0.067949 4.312 4.5783 1 {'Empirical'} 0.17621 0.0046171 -1.4103e-06 -0.0068855 0
E 0.00039896 0.00015673 9.4925e-05 0.00070697 0.9987 {'Empirical'} -0.00053724 -1.4103e-06 2.4564e-08 -1.8168e-05 0
WR 2.9792 0.16716 2.6506 3.3046 1 {'Empirical'} 0.11705 -0.0068855 -1.8168e-05 0.027943 0
Sigma2 10 0 10 10 1 {'Empirical'} 0 0 0 0 0
SummaryBeta contains the conditional posterior estimates.
Estimate the conditional posterior distributions of given that is the conditional posterior mean of (stored in SummaryBeta.Mean(1:(end – 1))). Return the estimation summary table.
condPostMeanBeta = SummaryBeta.Mean(1:(end - 1));
[~,SummarySigma2] = estimate(PriorMdl,X,y,'Beta',condPostMeanBeta);Method: lasso MCMC sampling with 10000 draws
Conditional variable: Beta fixed at -8.0643 4.4454 0.00039896 2.9792
Number of observations: 62
Number of predictors: 4
| Mean Std CI95 Positive Distribution
------------------------------------------------------------------------
Intercept | -8.0643 0.0000 [-8.064, -8.064] 0.000 Empirical
IPI | 4.4454 0.0000 [ 4.445, 4.445] 1.000 Empirical
E | 0.0004 0.0000 [ 0.000, 0.000] 1.000 Empirical
WR | 2.9792 0.0000 [ 2.979, 2.979] 1.000 Empirical
Sigma2 | 56.8314 10.2921 [39.947, 79.731] 1.000 Empirical
estimate displays an estimation summary of the conditional posterior distribution of given the data and that is condPostMeanBeta. In the display, inferences on are trivial.
Consider the regression model in Select Variables Using Bayesian Lasso Regression.
Create a prior model for performing SSVS. Assume that and are dependent (a conjugate mixture model). Specify the number of predictors p and the names of the regression coefficients.
p = 3; PriorMdl = mixconjugateblm(p,'VarNames',["IPI" "E" "WR"]);
Load the Nelson-Plosser data set. Create variables for the response and predictor series.
load Data_NelsonPlosser X = DataTable{:,PriorMdl.VarNames(2:end)}; y = DataTable{:,'GNPR'};
Implement SSVS by estimating the marginal posterior distributions of and . Because SSVS uses Markov chain Monte Carlo for estimation, set a random number seed to reproduce the results. Suppress the estimation display, but return the estimation summary table.
rng(1);
[PosteriorMdl,Summary] = estimate(PriorMdl,X,y,'Display',false);PosteriorMdl is an empiricalblm model object that stores draws from the posterior distributions of and given the data. Summary is a table with columns corresponding to posterior characteristics and rows corresponding to the coefficients (PosteriorMdl.VarNames) and disturbance variance (Sigma2).
Display the estimated parameter covariance matrix (Covariances) and proportion of times the algorithm includes each predictor (Regime).
Covariances = Summary(:,"Covariances")Covariances=5×1 table
Covariances
______________________________________________________________________
Intercept 103.74 1.0486 -0.0031629 0.6791 7.3916
IPI 1.0486 0.023815 -1.3637e-05 -0.030387 0.06611
E -0.0031629 -1.3637e-05 1.3481e-07 -8.8792e-05 -0.00025044
WR 0.6791 -0.030387 -8.8792e-05 0.13066 0.089039
Sigma2 7.3916 0.06611 -0.00025044 0.089039 74.911
Regime = Summary(:,"Regime")Regime=5×1 table
Regime
______
Intercept 0.8806
IPI 0.4545
E 0.0925
WR 0.1734
Sigma2 NaN
Regime contains the marginal posterior probability of variable inclusion ( for a variable). For example, the posterior probability that E should be included in the model is 0.0925.
Assuming that variables with Regime < 0.1 should be removed from the model, the results suggest that you can exclude the unemployment rate from the model.
Input Arguments
Bayesian linear regression model for predictor variable selection, specified as a model object in this table.
| Model Object | Description |
|---|---|
mixconjugateblm | Dependent, Gaussian-mixture-inverse-gamma conjugate model for SSVS
predictor variable selection, returned by bayeslm |
mixsemiconjugateblm | Independent, Gaussian-mixture-inverse-gamma semiconjugate model for SSVS
predictor variable selection, returned by bayeslm |
lassoblm | Bayesian lasso regression model returned by bayeslm |
Predictor data for the multiple linear regression model, specified as
a numObservations-by-PriorMdl.NumPredictors numeric
matrix. numObservations is the number of observations and must be
equal to the length of y.
Data Types: double
Response data for the multiple linear regression model, specified
as a numeric vector with numObservations elements.
Data Types: double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'Sigma2',2 specifies estimating the conditional posterior
distribution of the regression coefficients given the data and that the specified
disturbance variance is 2.
Flag to display Bayesian estimator summary to the command line, specified as the
comma-separated pair consisting of 'Display' and a value in this
table.
| Value | Description |
|---|---|
true | estimate prints estimation information and a
table summarizing the Bayesian estimators to the command line. |
false | estimate does not print to the command
line. |
The estimation information includes the estimation method, fixed parameters, the number of observations, and the number of predictors. The summary table contains estimated posterior means, standard deviations (square root of the posterior variance), 95% equitailed credible intervals, the posterior probability that the parameter is greater than 0, and a description of the posterior distribution (if known). For models that perform SSVS, the display table includes a column for variable-inclusion probabilities.
If you specify either Beta or Sigma2,
then estimate includes your specification in the display.
Corresponding posterior estimates are trivial.
Example: 'Display',false
Data Types: logical
Value of the regression coefficients for conditional posterior distribution estimation of the
disturbance variance, specified as the comma-separated pair consisting of
'Beta' and a (PriorMdl.Intercept +
PriorMdl.NumPredictors)-by-1 numeric vector.
estimate estimates the characteristics of
π(σ2|y,X,β
= Beta), where y is
y, X is X, and
Beta is the value of 'Beta'. If
PriorMdl.Intercept is true, then
Beta(1) corresponds to the model intercept. All other values
correspond to the predictor variables that compose the columns of
X. Beta cannot contain any
NaN values (that is, all coefficients must be known).
You cannot specify Beta and Sigma2 simultaneously.
By default, estimate does not compute characteristics of the conditional posterior of σ2.
Example: 'Beta',1:3
Data Types: double
Value of the disturbance variance for conditional posterior distribution estimation of the
regression coefficients, specified as the comma-separated pair
consisting of 'Sigma2' and a positive numeric
scalar. estimate estimates
characteristics of
π(β|y,X,Sigma2),
where y is y,
X is
X, and
Sigma2 is the value of
'Sigma2'.
You cannot specify Sigma2 and Beta
simultaneously.
By default, estimate does not compute characteristics of the conditional posterior of β.
Example: 'Sigma2',1
Data Types: double
Monte Carlo simulation adjusted sample size, specified as the comma-separated pair
consisting of 'NumDraws' and a positive integer.
estimate actually draws BurnIn –
NumDraws*Thin samples. Therefore,
estimate bases the estimates off NumDraws
samples. For details on how estimate reduces the full Monte
Carlo sample, see Algorithms.
Example: 'NumDraws',1e7
Data Types: double
Number of draws to remove from the beginning of the Monte Carlo sample to reduce transient
effects, specified as the comma-separated pair consisting of 'BurnIn'
and a nonnegative scalar. For details on how estimate reduces
the full Monte Carlo sample, see Algorithms.
Tip
To help you specify the appropriate burn-in period size, determine the extent of the transient behavior in the Monte Carlo sample by specifying 'BurnIn',0, simulating a few thousand observations using simulate, and then plotting the paths.
Example: 'BurnIn',0
Data Types: double
Monte Carlo adjusted sample size multiplier, specified as the comma-separated pair consisting of 'Thin' and a positive integer.
The actual Monte Carlo sample size is BurnIn + NumDraws*Thin. After discarding the burn-in, estimate discards every Thin – 1 draws, and then retains the next. For details on how estimate reduces the full Monte Carlo sample, see Algorithms.
Tip
To reduce potential large serial correlation in the Monte Carlo sample, or to reduce the
memory consumption of the draws stored in PosteriorMdl, specify
a large value for Thin.
Example: 'Thin',5
Data Types: double
Starting values of the regression coefficients for the Markov chain Monte Carlo (MCMC)
sample, specified as the comma-separated pair consisting of
'BetaStart' and a numeric column vector with
(PriorMdl.Intercept + PriorMdl.NumPredictors)
elements. By default, BetaStart is the ordinary least-squares (OLS)
estimate.
Tip
A good practice is to run estimate multiple times using
different parameter starting values. Verify that the solutions from each run
converge to similar values.
Example: 'BetaStart',[1; 2; 3]
Data Types: double
Starting values of the disturbance variance for the MCMC sample, specified as the
comma-separated pair consisting of 'Sigma2Start' and a positive
numeric scalar. By default, Sigma2Start is the OLS residual mean
squared error.
Tip
A good practice is to run estimate multiple times using different
parameter starting values. Verify that the solutions from each run converge to
similar values.
Example: 'Sigma2Start',4
Data Types: double
Output Arguments
Bayesian linear regression model storing distribution characteristics, returned as a
mixconjugateblm,
mixsemiconjugateblm, lassoblm, or empiricalblm model object.
If you do not specify either
BetaorSigma2(their values are[]), thenestimateupdates the prior model using the data likelihood to form the posterior distribution.PosteriorMdlcharacterizes the posterior distribution and is anempiricalblmmodel object. InformationPosteriorMdlstores or displays helps you decide whether predictor variables are important.If you specify either
BetaorSigma2, thenPosteriorMdlequalsPriorMdl(the two models are the same object storing the same property values).estimatedoes not update the prior model to form the posterior model. However,Summarystores conditional posterior estimates.
For more details on the display of PosteriorMdl, see
Summary.
Summary of Bayesian estimators, returned as a table. Summary
contains the same information as the display of the estimation summary
(Display). Rows correspond to parameters, and columns correspond
to these posterior characteristics:
Mean– Posterior meanStd– Posterior standard deviationCI95– 95% equitailed credible intervalPositive– Posterior probability that the parameter is greater than 0Distribution– Description of the marginal or conditional posterior distribution of the parameter, when knownCovariances– Estimated covariance matrix of the coefficients and disturbance varianceRegime– Variable-inclusion probabilities for models that perform SSVS; low probabilities indicate that the variable should be excluded from the model
Row names are the names in PriorMdl.VarNames. The name
of the last row is Sigma2.
Alternatively, pass PosteriorMdl to summarize to obtain a summary of Bayesian estimators.
More About
A Bayesian linear regression model treats the parameters β and σ2 in the multiple linear regression (MLR) model yt = xtβ + εt as random variables.
For times t = 1,...,T:
yt is the observed response.
xt is a 1-by-(p + 1) row vector of observed values of p predictors. To accommodate a model intercept, x1t = 1 for all t.
β is a (p + 1)-by-1 column vector of regression coefficients corresponding to the variables that compose the columns of xt.
εt is the random disturbance with a mean of zero and Cov(ε) = σ2IT×T, while ε is a T-by-1 vector containing all disturbances. These assumptions imply that the data likelihood is
ϕ(yt;xtβ,σ2) is the Gaussian probability density with mean xtβ and variance σ2 evaluated at yt;.
Before considering the data, you impose a joint prior distribution assumption on (β,σ2). In a Bayesian analysis, you update the distribution of the parameters by using information about the parameters obtained from the likelihood of the data. The result is the joint posterior distribution of (β,σ2) or the conditional posterior distributions of the parameters.
Tips
Monte Carlo simulation is subject to variation. If
estimateuses Monte Carlo simulation, then estimates and inferences might vary when you callestimatemultiple times under seemingly equivalent conditions. To reproduce estimation results, before callingestimate, set a random number seed by usingrng.
Algorithms
This figure shows how estimate reduces the Monte Carlo sample
using the values of NumDraws, Thin, and
BurnIn.
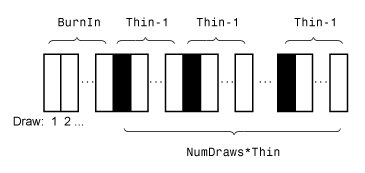
Rectangles represent successive draws from the distribution.
estimate removes the white rectangles from the Monte Carlo sample.
The remaining NumDraws black rectangles compose the Monte Carlo
sample.
Version History
Introduced in R2018b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)