dualtree2
Kingsbury Q-shift 2-D dual-tree complex wavelet transform
Description
[
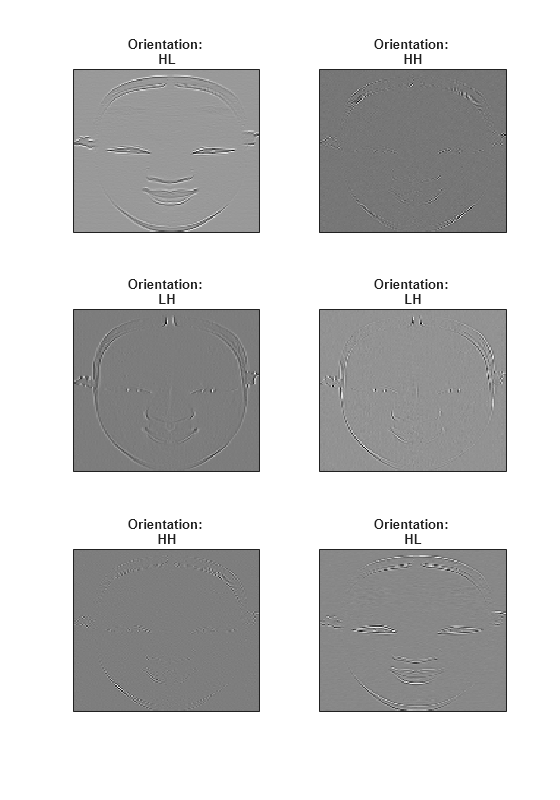
returns the 2-D dual-tree complex wavelet transform (DTCWT) of A,D] = dualtree2(X)X using
Kingsbury Q-shift filters. The output A is the matrix of real-valued
final-level scaling (lowpass) coefficients. The output D is a
L-by-1 cell array of complex-valued wavelet coefficients, where
L is the level of the transform. For each element of
D there are six wavelet subbands.
The DTCWT is obtained by default down to level
floor(log2(min([H
W]))), where H and W
refer to the height (row dimension) and width (column dimension) of X,
respectively. If any of the row or column dimensions of X are odd,
X is extended along that dimension by reflecting around the last row
or column.
By default, dualtree2 uses the near-symmetric biorthogonal wavelet
filter pair with lengths 5 (scaling filter) and 7 (wavelet filter) for level 1 and the
orthogonal Q-shift Hilbert wavelet filter pair of length 10 for levels greater than or equal
to 2.
[___] = dualtree2(
specifies additional options using name-value pair arguments. For example,
X,Name,Value)'LevelOneFilter','antonini' specifies the (9,7)-tap Antonini filter as
the biorthogonal filter to use in the first-level analysis.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Antonini, M., M. Barlaud, P. Mathieu, and I. Daubechies. “Image Coding Using Wavelet Transform.” IEEE Transactions on Image Processing 1, no. 2 (April 1992): 205–20. https://doi.org/10.1109/83.136597.
[2] Kingsbury, Nick. “Complex Wavelets for Shift Invariant Analysis and Filtering of Signals.” Applied and Computational Harmonic Analysis 10, no. 3 (May 2001): 234–53. https://doi.org/10.1006/acha.2000.0343.
[3] Le Gall, D., and A. Tabatabai. “Sub-Band Coding of Digital Images Using Symmetric Short Kernel Filters and Arithmetic Coding Techniques.” In ICASSP-88., International Conference on Acoustics, Speech, and Signal Processing, 761–64. New York, NY, USA: IEEE, 1988. https://doi.org/10.1109/ICASSP.1988.196696.
Extended Capabilities
Version History
Introduced in R2020a